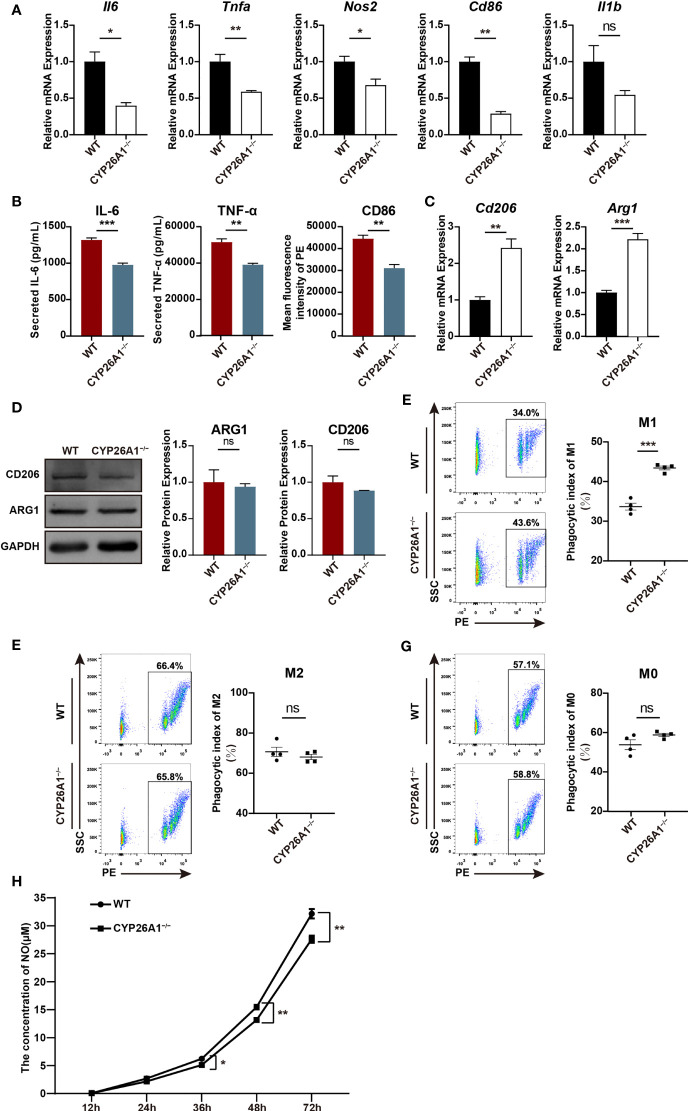
Figure 5.
CYP26A1 deficiency affected the polarization, phagocytic capacity and NO production capacity of Raw264.7 cells. (A) qPCR analysis of M1 phenotype genes Il6, Nos2, Tnfa, Cd86 and Il1b in CYP26A1-deficient Raw264.7 and WT cells after induced 4 h by LPS and IFN-γ (n=3). (B) ELISA was used to measure IL-6 and TNF-α secretion in supernatants of Raw264.7 cells (WT and CYP26A1−/−) treated with LPS and IFN-γ for 24 h; Mean fluorescence intensity analysis of CD86 expression levels in Raw264.7 cells (WT and CYP26A1−/−) incubation with LPS and IFN-γ for 24 h (n=3). (C) qPCR analysis of M2 markers Arg1 and Cd206 in CYP26A1 knockout Raw264.7 and WT group after induced 4 h by IL-4 and IL-13 (n=3). (D) Western blot analysis of M2 markers ARG1 and CD206 in CYP26A1 knockout Raw264.7 and WT group after induced 24 h by IL-4 and IL-13 (n=3). GAPDH was used as loading control. (E–G) Flow cytometry analysis of the phagocytic capacity of Raw264.7 (WT and CYP26A1−/−) under M0, M1 and M2 state (n=4). (H) The concentration of NO in the supernatant of RAW264.7 cells (WT and CYP26A1−/−) treated with LPS and IFN-γ at different induction time (12 h, 24 h, 36 h, 48 h, 72 h) was detected by griess reagent (n=4). Error bars represent means ± SEM; two-tailed unpaired t-test, ns, not significant, *P < 0.05, **P < 0.01, or ***P < 0.001.