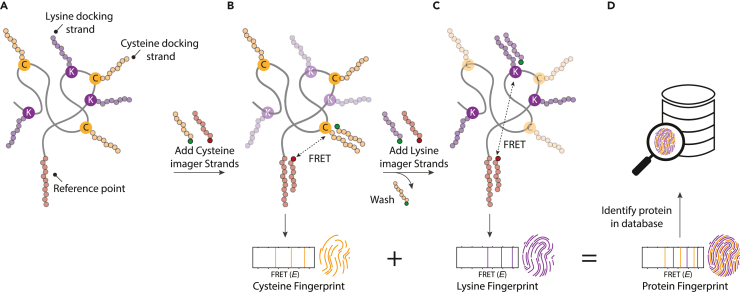
Figure 1.
The concept of FRET X for protein fingerprinting
(A) A subset of amino acids (here cysteines and lysines) are labeled with orthogonal DNA sequences that function as docking sites for complementary, fluorescently labeled imager strands. Another orthogonal DNA sequence is conjugated to one of the protein termini, which serves as an acceptor docking site and facilitates immobilization of the protein to a microfluidic device.
(B) In the first round of FRET X imaging, imager strands that hybridize with the cysteine docking site (yellow circles) and those that hybridize with the reference point (red circles) are injected in the microfluidic chamber. Both the donor and acceptor-labeled imager strands transiently interact with their complementary docking strands. When both are present at the same time, FRET can occur and the FRET efficiency is determined between a cysteine and the reference point. Each of the three FRET pairs is separately probed, giving rise to a number of FRET efficiencies (E), which constitute the cysteine fingerprint.
(C) The chamber is washed and FRET X imaging is repeated to probe the lysines. This FRET X cycle can be repeated to probe additional amino acids and generate additional fingerprints.
(D) The FRET efficiencies for individual amino acids are combined to produce a protein fingerprint that can be mapped against a reference database to identify the protein.