Figure 2.
Model peptides can be fingerprinted with FRET X
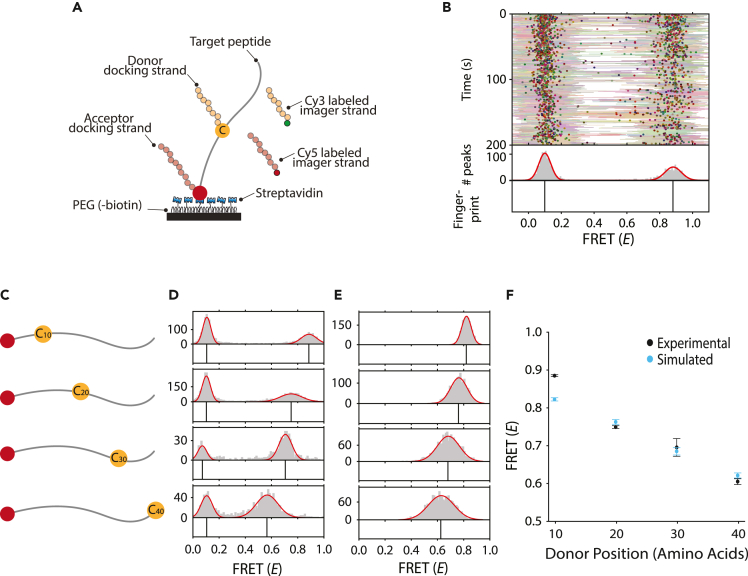
(A) Depiction of the experimental system for peptide fingerprinting. The target peptide is immobilized through conjugation of its N-terminal biotin with the streptavidin on the PEGylated surface. The donor (Cy3)-labeled imager strand (yellow) can bind to the DNA-docking site on the cysteine, while the acceptor (Cy5)-labeled imager strand (red) can hybridize to the docking site on the lysine. Simultaneous binding generates short FRET events and is observed with total internal reflection microscopy.
(B) Representative kymograph for a peptide with a cysteine that is 10 amino acids separated from the acceptor-binding site. The FRET efficiency for each data point in a binding event (lines) and the mean FRET efficiency from all data points in a binding event (dots) are indicated as a function of time. A Gaussian distribution (0.88 ± 0.14) is fitted on a histogram of average FRET efficiencies per FRET event. The means of the Gaussians are plotted in a separate panel (bottom) and are referred to as the FRET X fingerprint of the peptide. The FRET population on the left is caused by donor leakage into the acceptor channel.
(C) Our four model peptides have a lysine at the N terminus and a cysteine at position 10, 20, 30 or 40. See Table S1 for the full amino acid sequences of the model peptides.
(D) Experimental distributions and fingerprints for each peptide show a downward trend in mean FRET (E) for increasing FRET pair separation (mean ± FWHM of the Gaussian fit: 0.89 ± 0.14, 0.75 ± 0.20, 0.72 ± 0.11, 0.57 ± 0.20). See also Figures S2 and S3 for imager strand dwell times and kymographs for single peptides, respectively.
(E) The simulated distributions and fingerprints for the four peptides show a similar downward trend in distribution means (0.82 ± 0.08, 0.76 ± 0.15, 0.68 ± 0.20, 0.62 ± 0.23).
(F) Experimental and simulated data correlate well. Whiskers denote ± one standard deviation. Standard deviation of experimental data points is over four kymographs (each consisting of hundreds of events). Experiments were performed on separate days.