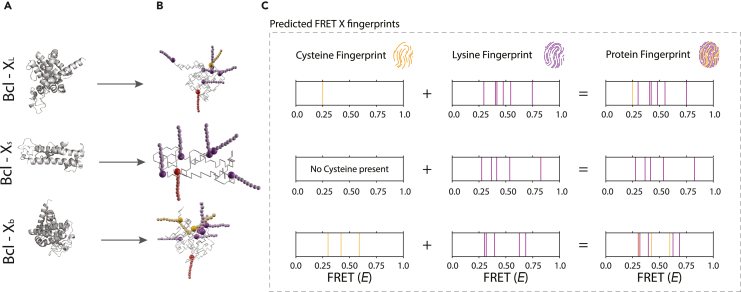
Figure 3.
Representative FRET (E) fingerprints for three spliceoforms of BCL-X
(A) Fully atomic structure for BCL XL, Xs, and Xb (from top to bottom) as predicted by the RaptorX structure prediction tool (Källberg et al., 2012, 2014).
(B) Energy-optimized lattice model structures with DNA-docking strands attached to cysteines (orange) and lysines (purple). The reference acceptor docking strand (red) is added to the N terminus of the proteins.
(C) The simulated fingerprint for spliceoform of the BCL proteins. Fingerprints are based on averaged donor-acceptor distances in 100 structural snapshots of Markov chain-generated lattice model structures (distributions shown in Figure S4). Fingerprints for a second set of spliceoforms (PTGS1) are shown in Figure S5.