Abstract
Antibodies to the nucleocapsid (N) antigen are suggested to be used to monitor infections after COVID-19 vaccination, as first generation subunit vaccines are based on the spike (S) protein. We used multiplex immunoassays to simultaneously measure antibody responses to different fragments of the SARS-CoV-2 S and N antigens for evaluating the immunogenicity of the mRNA-1273 (Spykevax) and the BNT162b2 (Comirnaty) vaccines in 445 health care workers. We report a >4-fold increase post-vaccination of IgG levels to the full length (N FL) and C-terminus of N (N CT) in 5.2% and 18.0% of individuals, respectively, and of IgA in 3.6% (N FL) and 9.0% (N CT) of them. The increase in IgG levels and avidity was more pronounced after Spykevax than Comirnaty vaccination (36.2% vs 13.1% for N CT, and 10.6% vs 3.7% for N FL). Data suggest the induction of cross-reactive antibodies against the N CT region after administering these S-based vaccines, and this should be taken into account when using N seropositivity to detect breakthroughs.
Abbreviations: spike, S; nucleocapsid, N; C-terminal, CT; full length, FL; receptor-binding domain, RBD; median fluorescence intensity, MFI; standard deviations, SD; confidence intervals, CI; interquartile range, IQR; health care workers, HCW; fold change, FC; N-terminal, NT
At A Glance Commentary.
Dobaño C, et al.
Background
Longitudinal studies in cohorts of health care workers including naïve and SARS-CoV-2 pre-exposed individuals allow monitoring the antibody responses to vaccination and infection. We report the unexpected finding that immunization with the mRNA-1273 (Spykevax) and the BNT162b2 (Comirnaty) vaccines can lead to a significant increase in antibodies to the nucleocapsid (N) protein, particularly the C-terminus fragment.
Translational Significance
This is relevant because this N antigen, which is not included in the immunogen, is suggested to monitor vaccine breakthroughs. A potential cross-reactivity would lead to a overestimation of vaccine failure, therefore alternative antigens need to be used to evaluate vacine effectiveness as part of the global implementation of COVID-19 immunization.
Alt-text: Unlabelled box
INTRODUCTION
With the successful completion of phase 3 clinical trials1 and subsequent global rollout of COVID-19 vaccines since December 2020, there is a need to monitor the occurrence of breakthroughs and the duration of protection, for the evaluation of the effectiveness of immunization. This is becoming of paramount importance with the appearance of new more transmissible variants that may potentially escape immune responses elicited by COVID-19 vaccines. Indeed, effectiveness of COVID-19 vaccines may be lower to the Delta variant.2 As first generation subunit vaccines are based on the spike (S) protein from SARS-CoV-2,3 infections in vaccinated individuals using serological testing cannot be detected with antibodies to this target. Therefore, current studies suggest using antibodies to the nucleocapsid (N) antigen to diagnose incident infections after vaccination.4 , 5
As part of a longitudinal evaluation of the immunogenicity of the mRNA-1273 (Spykevax)1 and the BNT162b2 (Comirnaty) vaccines in a cohort of health care workers followed up since the onset of the COVID-19 pandemic in 2020, we set out to monitor IgM, IgA and IgG responses to several SARS-CoV-2 antigens including S and N proteins after vaccination started in early 2021.
METHODS
Study population
We followed up a total of 445 health care workers (HCW, n = 353 from hospital6 and n = 92 from primary care7) in Barcelona, Spain, with different histories of prior COVID-19 exposure. Venous or finger prick blood was collected at different time points before and after immunization with the mRNA-1273 (Spykevax)1 or the BNT162b2 (Comirnaty),8 products from Moderna Biotech and Pfizer-BioNTech, respectively. Plasma was isolated and cryopreserved at -80°C.
Antibody determinations
We used a multiplex immunoassay able to simultaneously measure levels of antibodies to different fragments of the S and N proteins. The N antigens included a C-terminal (CT) fragment (residues 340-416) and the full length (FL) of N, both cloned in pET22b expression vectors, fusing a C-terminal 6xHis-tag, transformed in E. coli BL21 DE3, induced with IPTG, and purified by affinity chromatography using HisTrap columns. The integrity of the proteins was checked by Coomassie stained SDS-PAGE, showing >90% purity.9 , 11 The S antigens included a full length protein, as well as its subregion S2, and the receptor-binding domain (RBD), as previously reported.10 , 11 Antigen-coupled microspheres were added to 384 well plates in multiplex and incubated with test plasma samples, positive and negative controls (1:500 final dilution) for 1 hour at room temperature, protected from light. After washing, goat anti-human IgG (1:400) or IgA (1:200) phycoerythrin conjugates (Moss Bio) were added to each well and incubated for 30 minutes. After washing, plates were acquired on a Flexmap 3D reader, at least 50 microspheres per analyte per well, and median fluorescence intensity (MFI) reported for each analyte. Isotype and antigen specific seropositivity cutoffs were calculated as 10 to the mean plus 3 standard deviations (SD) of log10-transformed MFI of 128 pre-pandemic negative control samples.
Samples from a subset of 82 individuals (41 vaccinated with Comirnaty [5 pre-exposed] and 41 with Spykevax vaccine [14 pre-exposed]), were selected for avidity assays. Antibody avidity was determined as the percentage of IgG and IgA levels against S, N FL and N CT antigens measured incubating plasma samples (diluted 1:500) with a chaotropic agent (urea 4M, 30 min at room temperature) over the IgG or IgA levels measured in the same samples without chaotropic agent.
Statistical analysis
MFIs were log10-transformed. Groups were compared with the Wilcoxon Sum Rank test for continuous non-parametric variables and with the Wilcoxon Signed-Sum Rank Test for paired non-parametric continuous data. Correlations between continuous variables were analyzed using linear regression models and Spearman's rank test. A P-value of < 0.05 was considered statistically significant and 95% confidence intervals (CI) were calculated for all estimates. We performed the statistical analysis using GraphPad Prism V9.
Ethics
The study protocol was approved by the Comitè Ètic d'Investigació Clínica IDIAP Jordi Gol and the CEIm of Hospital Clínic, and written informed consent was obtained from all participants.
RESULTS
While responses to S proteins were generally induced at high levels pre- to post-vaccination, as expected, we observed an unanticipated statistically significant increase in antibody responses to N proteins, particularly against the N CT fragment (Fig 1 A). The rise in antibody levels for each antigen was consistent with an increase in percent seropositivity (Table I). As predictable, most naïve subjects were seronegative for anti-N antibodies prior to vaccination.
Fig 1.
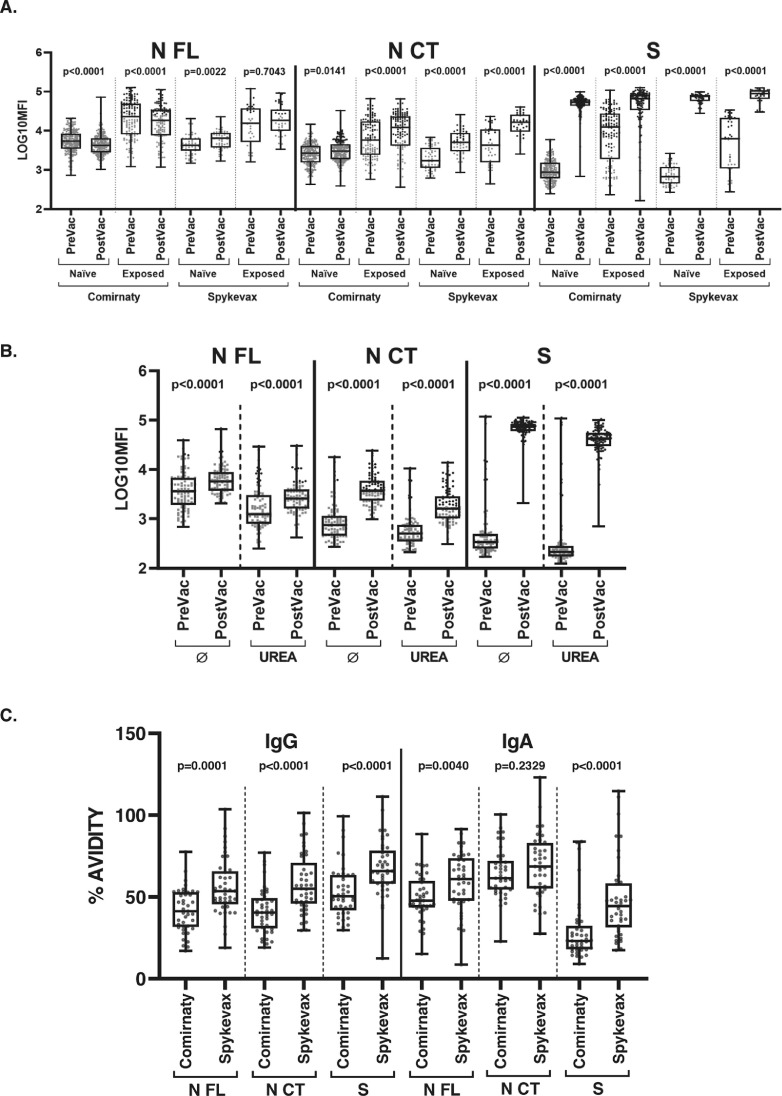
Comparison of IgG levels to N FL, N CT and S, before and after COVID-19 mRNA vaccination, and IgG and IgA avidity post-vaccination. IgG levels (log10 MFI) against the N FL, N CT and S proteins of SARS-CoV-2 before and after COVID-19 vaccination stratifying by vaccine and prior SARS-CoV-2 exposure status (A); IgG levels comparing with and without urea incubation (B); and IgG and IgA % of avidity post-vaccination stratifying by vaccine (C). The center line of boxes depicts the median of MFIs; the lower and upper hinges correspond to the first and third quartiles; the distance between the first and third quartiles corresponds to the interquartile range (IQR); whiskers extend from the hinge to the highest or lowest value within 1.5 × IQR of the respective hinge. Black dots correspond to seropositive samples and grey dots correspond to seronegative samples. Wilcoxon rank test was used to assess statistically significant differences in antibody levels between paired samples.
Table I.
Antibody seropositivity in health care personnel before and after COVID-19 mRNA vaccination, and percentage of subjects with a >4-fold increase in antibody levels, stratifying by antibody isotype, antigen, prior SARS-CoV-2 exposure status and vaccine.
| Naïve |
Exposed* |
|||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Comirnaty |
Spykevax |
Comirnaty |
Spykevax |
|||||||||||||||||||||||||
| PreVac | PostVac | FC >4 | PreVac | PostVac | FC >4 | PreVac | PostVac | FC >4 | PreVac | PostVac | FC >4 | |||||||||||||||||
| IgG | N CT | 0.0% | (0) | 21.3% | (48) | 9.8% | (22) | 0.0% | (0) | 50.9% | (28) | 29.1% | (16) | 42.9% | (54) | 67.5% | (85) | 19.0% | (24) | 38.5% | (15) | 94.9% | (37) | 46.2% | (18) | |||
| N FL | 0.0% | (0) | 0.9% | (2) | 4.0% | (9) | 0.0% | (0) | 0.0% | (0) | 3.6% | (2) | 46.0% | (58) | 38.9% | (49) | 3.2% | (4) | 41.0% | (16) | 41.0% | (16) | 20.5% | (8) | ||||
| RBD | 0.0% | (0) | 99.6% | (224) | 99.6% | (224) | 1.8% | (1) | 100.0% | (55) | 100.0% | (55) | 69.0% | (87) | 94.4% | (119) | 62.7% | (79) | 59.0% | (23) | 100.0% | (39) | 92.3% | (36) | ||||
| S | 0.0% | (0) | 99.6% | (224) | 99.6% | (224) | 1.8% | (1) | 100.0% | (55) | 100.0% | (55) | 70.6% | (89) | 92.9% | (117) | 51.6% | (65) | 56.4% | (22) | 100.0% | (39) | 76.9% | (30) | ||||
| S2 | 0.9% | (2) | 97.3% | (219) | 93.3% | (210) | 0.0% | (0) | 100.0% | (55) | 100.0% | (55) | 68.3% | (86) | 85.7% | (108) | 23.8% | (30) | 56.4% | (22) | 100.0% | (39) | 46.2% | (18) | ||||
| IgA | N CT | 1.3% | (3) | 2.7% | (6) | 0.4% | (1) | 0.0% | (0) | 14.5% | (8) | 7.3% | (4) | 22.2% | (28) | 69.0% | (87) | 23.8% | (30) | 5.1% | (2) | 46.2% | (18) | 12.8% | (5) | |||
| N FL | 1.8% | (4) | 0.4% | (1) | 0.4% | (1) | 1.8% | (1) | 1.8% | (1) | 1.8% | (1) | 16.7% | (21) | 34.9% | (44) | 11.1% | (14) | 12.8% | (5) | 7.7% | (3) | 0.0% | (0) | ||||
| RBD | 0.0% | (0) | 96.0% | (216) | 79.6% | (179) | 0.0% | (0) | 98.2% | (54) | 94.5% | (52) | 47.6% | (60) | 95.2% | (120) | 77.0% | (97) | 35.9% | (14) | 100.0% | (39) | 84.6% | (33) | ||||
| S | 0.4% | (1) | 98.2% | (221) | 94.2% | (212) | 0.0% | (0) | 98.2% | (54) | 98.2% | (54) | 57.1% | (72) | 96.0% | (121) | 84.9% | (107) | 46.2% | (18) | 100.0% | (39) | 92.3% | (36) | ||||
| S2 | 1.3% | (3) | 84.4% | (190) | 85.8% | (193) | 0.0% | (0) | 87.3% | (48) | 94.5% | (52) | 52.4% | (66) | 92.9% | (117) | 65.1% | (82) | 38.5% | (15) | 94.9% | (37) | 76.9% | (30) | ||||
| Number subjects | 225 | 55 | 126 | 39 | ||||||||||||||||||||||||
Abbreviations: FC, fold change; N, nucleocapsid; RBD, receptor binding domain; S, spike.
There were 39 (8.7%) and 126 (28.3%) SARS-CoV-2 pre-exposed participants among those vaccinated with Spykevax (Moderna BioTech) and with Comirnaty (Pfizer-BioNTech), respectively, defined by positive SARS-CoV-2 rRT-PCR and/or serology diagnosis before vaccination in the prospective cohorts of health care workers (HCW). Antibody levels were measured in the prior visit of the longitudinal follow up that was available pre-immunization (PreVac), depending on the study cohort (ranging from 1 day to 6 months), and post-immunization (PostVac) at 5 to 34 days after the 2nd dose (except for 77 HCW samples measured 3 to 21 days post 1st dose). Percentages of seropositivity are shown (%), and in parenthesis the number of individuals, over the total of paired samples shown in the bottom row.
Specifically, levels of IgG and IgA, to the N CT fragment rose >4-fold in 18.06% and 9.0% of vaccinated individuals, respectively (Table S1). This increase was not observed for IgM (data not shown). Remarkably, the 4-fold increase in IgG was observed in more individuals for the Spykevax (36.2%) than the Comirnaty (13.1%) vaccination (Tables 1, S1) and the increase in IgG reached more statistical significance for the Moderna product (P < 0.001) (Fig 1A).
Examining the antibody responses to N FL, commonly used in serological evaluations, 5.2% and 3.6% of individuals had a >4-fold increase in IgG and IgA levels, respectively (Table S1), but the patterns depended on the vaccine. Following COVID-19 Spykevax vaccination, levels of IgG and IgA to N FL increased but to a lesser extent than to N CT (Fig 1A), with 10.6% of individuals showing a >4-fold change in IgG levels (Table S1). In contrast, N FL IgG levels decreased pre- to post-vaccination with Comirnaty in most of the participants (Fig 1A).
Correlations between levels of antibodies to N CT and S proteins (full length S, S2, RBD) post-vaccination were significant (Spearman rho, 0.70-0.83 for IgG, and 0.68-0.88 for IgA, P < 0.0001), and a similar but weaker correlation was observed between levels of antibodies to N FL and S proteins post-vaccination (Spearman rho, 0.50-0.76 for IgG, and 0.53-0.85 for IgA, P < 0.0001). Correlations were also significant but less strong when analyzing pre-vaccination antibody levels: N CT, rho 0.51-0.72 for IgG, and 0.44-0.75 for IgA, P < 0.0001; N FL, rho 0.31-0.53 for IgG, and 0.36-0.67 for IgA, P < 0.0001.
We performed additional experiments to understand the observed raise in antibodies to an antigen (N) that is not included in the vaccine. We first discarded non-specific effects of the mRNA COVID-19 vaccines, or bystander activity, since antibody responses to other vaccine antigens (hepatitis B virus, diphtheria, tetanus, pertussis) or to N from common cold human coronaviruses did not significantly increase; in fact, some of them decreased after vaccination like N FL (data not shown). We also discarded that this antibody increase was due to multispecific or polyreactive memory T helper or B cells in SARS-CoV-2 pre-exposed individuals, whereby a booster response against S through vaccination would also recall responses against non-vaccine antigens in the virus (N), since this phenomenon was also observed in naïve volunteers. The finding that pre-exposed individuals had a stronger increase in N CT IgG levels after vaccination mirrors the booster effect that is seen with S (Fig 1A).
Since correlation analyses suggested that antibodies induced by both mRNA vaccines could be due to cross-reactivity between N and S epitopes, we aligned the primary amino acid sequences of N CT (residues 340-416)9 and S but found a negligible identity (1.5%) in the overall sequence. A short C-terminal stretch in S1 (upstream of RBD), between amino acids 179 and 256, showed a 13% of identity with N CT, similar to that observed between specific immunodominant regions of common cold human coronaviruses with minimal cross-reactivity with the same N CT fragment.9 Although these alignments did not provide strong evidence for cross-reactivity, at least at the linear epitope level, we also tested other fragments of the N protein to try mapping the target epitopes. Different fragments of N based on the N-terminal domain (NT, residues 43-180), C-terminal domain (residues 250-360) or shorter peptides within the C-terminal domain (residues 384-406), were not recognized by antibodies elicited upon vaccination in this cohort (Figure S2 and data not shown).
Finally, testing the avidity of antibodies following vaccination, we found that cross-reactive antibodies did not bind weakly to N (Fig 1B). Thus, the increase in anti-N antibody levels pre- to post-vaccination remained statistically significant when incubating samples with a chaotropic agent. The strength of IgG antibodies binding N CT was only slightly lower than that of IgG binding S, while IgA to N CT had higher avidity than IgA to S (Fig 1C). Spykevax vaccination induced statistically significantly higher avidity S and N CT antibodies than Comirnaty vaccination.
DISCUSSION
We report an increase in IgG and IgA to the N protein, specifically to the N CT, early after COVID-19 vaccination, with a more pronounced IgG rise for the Spykevax than the Comirnaty vaccine. These results suggest the presence of cross-reactive antibodies against the C-terminal region of the SARS-CoV-2 N protein after vaccination with S-based mRNA vaccines. Avidity assays indicate that the binding of vaccine-induced cross-reactive antibodies to N CT epitopes is not mediated by weak interactions that are characteristic of non-specific or polyreactive antibodies. We hypothesize that the amino acid residues recognized by these antibodies are either constituted by conformational epitopes,13 or very short amino acid stretches not captured by sequence alignment, possibly located between amino acids 340-416 of N, considering that B cell epitopes can be as short as 4-5 amino acids in length.
A lack of antibody response against N FL after vaccination with mRNA vaccines has been previously reported,5 , 12 and the decay we observe is consistent with a gradual waning of N antibodies with time. In kinetics studies, antibodies to N FL tend to decrease more markedly and rapidly with time post-infection compared to S antibodies. Therefore, the pattern found here could reflect the dynamics of the IgG response to N FL after infection with SARS-CoV-2 (in exposed) and/or to common cold human coronaviruses (in naive), which keep decreasing from pre to post vaccination. Besides the natural antibody decay, the decrease in IgG to N FL upon vaccination could be related to a redirection of antibody production to vaccine specific immunogens in detriment of other targets.
Finally, it is highly unlikely that the increase in IgG and IgA levels against N post-vaccination is due to asymptomatic infections given the high proportion of antibody increasers. Moreover, upon recent infection, the magnitude of the increase in N FL antibodies would be much bigger particularly in previously exposed individuals, thus this option was also discarded.
Our results have significant public health relevance because the N protein is being suggested to detect breakthroughs in vaccinated people.4 Our findings included HCW from separate cohorts, with serology assays performed in independent experiments, with antibodies measured at different time-points pre- and post-vaccination, and showing consistent results. Therefore, if this considerable number of individuals has cross-reactive antibodies between S and N CT, by using seropositivity to N as a marker of infection, they would falsely be classified as infected, and this would affect the accuracy of vaccine effectiveness estimates. The rise in antibodies is less marked for N FL, as the polyclonal response is the summation of different epitope specificities, including N CT but also N NT and central regions that were not affected by vaccination. Thus, the cross-reactive antibodies reaching the N CT contained in the N FL are proportionally less compared to the N CT epitopes alone, giving lower levels of cross-reactivity. The impact of false positive SARS-CoV-2 infections appears to occur more frequently with the COVID-19 Moderna vaccine, with which millions of people are being vaccinated. The higher increase in anti-N CT IgG in the Moderna vaccine recipients vs the Pfizer could be the result from higher levels of anti-S antibodies being induced by the Moderna vaccine.14 In fact, the proportion of vaccine breakthrough infections in the USA15 appears lower than the proportion of vaccinated individuals showing an increase in anti-N antibodies in our study.
In conclusion, in light of these findings, increase in antibody levels to N in vaccinated individuals may not be the best strategy to detect viral breakthroughs, particularly if using N CT fragments, although N FL seropositivity appears to be adequate. The World Health Organization indicates that careful consideration should be made as to which antibody target is selected, and alludes to the faster waning of anti-N protein antibodies after infection.16 Therefore, combinations of multiple antigens including other less studied non-structural proteins that have been shown to be immunogenic17 have greater potential to accurately discriminate signatures of vaccine failure, for a more robust evaluation of vaccine effectiveness.
ACKNOWLEDGMENTS
We thank the volunteers and research teams participating in the SeroCov1 and CovidCatCentral studies; Marta Tortajada, Sonia Barroso, Gemma Salmerón, Anna Llupià and Anna Vilella, from the clinical teams; Mª José Molina, Diana Barrios and Esther Prados, from the ISGlobal laboratory team; Carlo Carolis, Natalia Rodrigo Melero, Jordi Chi, Laia Fernández-Barat and Rubén López, for procurement of N antigens; Gemma Ruiz-Olalla, Susana Méndez, Pau Rodó, Natalia Ortega, and Marta Ribes, for database management; Cristina Castellana for project assistance.
The study received support from Fundació Privada Daniel Bravo Andreu, ISGlobal internal funds, in-kind contributions of Hospital Clínic de Barcelona, and the European Institute of Innovation and Technology (EIT) Health (grant number 20877), supported by the European Institute of Innovation and Technology, a body of the European Union receiving support from the H2020 Research and Innovation Programme. L. I. work was supported by PID2019-110810RB-I00 grant from the Spanish Ministry of Science & Innovation. Development of SARS-CoV-2 reagents was partially supported by the National Institute of Allergy and Infectious Diseases Centers of Excellence for Influenza Research and Surveillance (contract number HHSN272201400008C). ISGlobal is supported by the Ministerio de Ciencia e Innovación through the Centro de Excelencia Severo Ochoa 2019-2023 program (grant number CEX2018-000806-S), and the Catalan Government through the Centres de Recerca de Catalunya program (CERCA).
All authors have read the journal's policy on disclosure of potential conflicts of interest and the journal's authorship statement. The manuscript has been reviewed and approved by all authors.
The authors declare that no conflict of interests exist.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.trsl.2021.10.004.
Appendix. Supplementary materials
REFERENCES
- 1.Baden LR, El Sahly HM, Essink B, et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N Engl J Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kislaya I, Rodrigues EF, Borges V, et al. Delta variant and mRNA Covid-19 vaccines effectiveness: higher odds of vaccine infection breakthroughs. MedRxiv. 2021 doi: 10.1101/2021.08.14.21262020. [DOI] [Google Scholar]
- 3.Dai L, Gao GF. Viral targets for vaccines against COVID-19. Nat Rev Immunol. 2021;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dörschug A, Frickmann H, Schwanbeck J, et al. Comparative Assessment of Sera from Individuals after S-Gene RNA-Based SARS-CoV-2 Vaccination with Spike-Protein-Based and Nucleocapsid-Based Serological Assays. Diagnostics. 2021;11:426. doi: 10.3390/diagnostics11030426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Assis R, Jain A, Nakajima R, et al. Distinct SARS-CoV-2 antibody reactivity patterns elicited by natural infection and mRNA vaccination. NPJ Vaccines. 2021;6(1):132. doi: 10.1038/s41541-021-00396-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Basteiro AL, Moncunill G, Tortajada M, et al. Seroprevalence of antibodies against SARS-CoV-2 among health care workers in a large Spanish reference hospital. Nat Commun. 2020;11:3500. doi: 10.1038/s41467-020-17318-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobaño C, Ramírez-Morros A, Alonso S, et al. Persistence and baseline determinants of seropositivity and reinfection rates in health care workers up to 12.5 months after COVID-19. BMC Med. 2021;19:155. doi: 10.1186/s12916-021-02032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polack FP, Thomas SJ, Kitchin N, et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobaño C, Santano R, Jiménez A, et al. Immunogenicity and crossreactivity of antibodies to the nucleocapsid protein of SARS-CoV-2: utility and limitations in seroprevalence and immunity studies. Transl Res. 2021;21 doi: 10.1016/j.trsl.2021.02.006. S1931-5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amanat F, Nguyen T, Chromikova V, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020 doi: 10.1101/2020.03.17.20037713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobaño C, Vidal M, Santano R, et al. Highly sensitive and specific multiplex antibody assays to quantify immunoglobulins M, A and G against SARS-CoV-2 antigens. J Clin Microbiol. 2020;59 doi: 10.1101/2020.06.11.147363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradley T, Grundberg E, Selvarangan R, et al. Antibody Responses after a Single Dose of SARS-CoV-2 mRNA Vaccine. N Engl J Med. 2021;384:1959–1961. doi: 10.1056/NEJMc2102051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Negi SS, Braun W. Cross-React: a new structural bioinformatics method for predicting allergen cross-reactivity. Bioinformatics. 2017;33:1014–1020. doi: 10.1093/bioinformatics/btw767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steensels D, Pierlet N, Penders J, Mesotten D, Heylen L. Comparison of SARS-CoV-2 Antibody Response Following Vaccination With BNT162b2 and mRNA-1273. JAMA. 2021 doi: 10.1001/jama.2021.15125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CDC. COVID-19 Vaccine Breakthrough Case Investigation and Reporting. CDC; COVID Vaccination; Heal 2021: Available at: https://www.cdc.gov/vaccines/covid-19/health-depar.
- 16.WHO. Evaluation of COVID-19 vaccine effectiveness: interim guidance, 2021. World Heal Organ 2021:viii, 58 p.
- 17.DiMuzio JM, Heimbach BC, Howanski RJ, et al. Unbiased interrogation of memory B cells from convalescent COVID-19 patients reveals a broad antiviral humoral response targeting SARS-CoV-2 antigens beyond the spike protein. Vaccine X. 2021;8 doi: 10.1016/j.jvacx.2021.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.