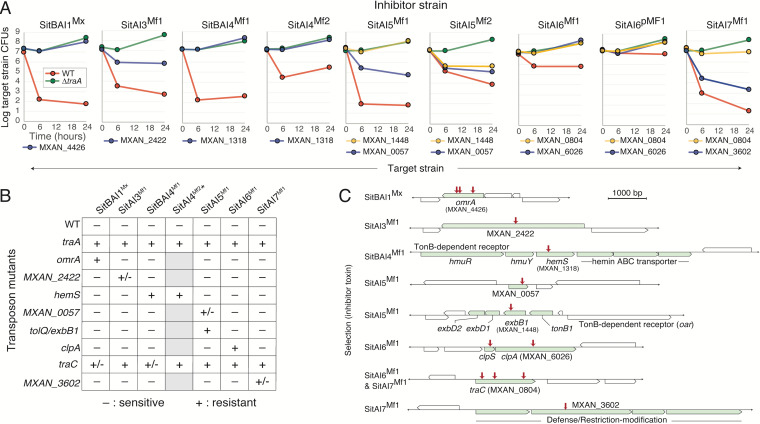
FIG 2.
Resistance phenotypes and gene identities of SitA-resistant mutants. (A) Competition assays determined by CFU of backcrossed mutants (Kanr) against inhibitor strains expressing the indicated sitAI alleles (Kans) at various times. Sensitive (WT) (red lines) and resistant (traA::Kmr) (green lines) controls are shown. Locus tags denote genes with mini-Himar insertions. For sitA4, -5, and -6, additional toxin alleles were used to test for cross-resistance within a family. (B) Cross-resistance assessment of inhibitors against target strains. Assays were done as described above for panel A, where a single 24-h time point was measured. −, sensitive; +, resistant (within 1 log of the traA mutant at 24 h); +/−, partial resistance (between sensitive and resistant controls). Note that the presence of the sitB gene in the indicated inhibitor strains (top) correlated with limited or partial resistance by the traC mutant. *This native operon does not contain a sitB gene and was not used in the screen (Fig. 1). Gray boxes, not tested. (C) Genes identified from the screen, their neighborhoods, and the inhibitors that they were isolated against. Red arrows represent insertion sites. See Table S1A in the supplemental material for strain details.