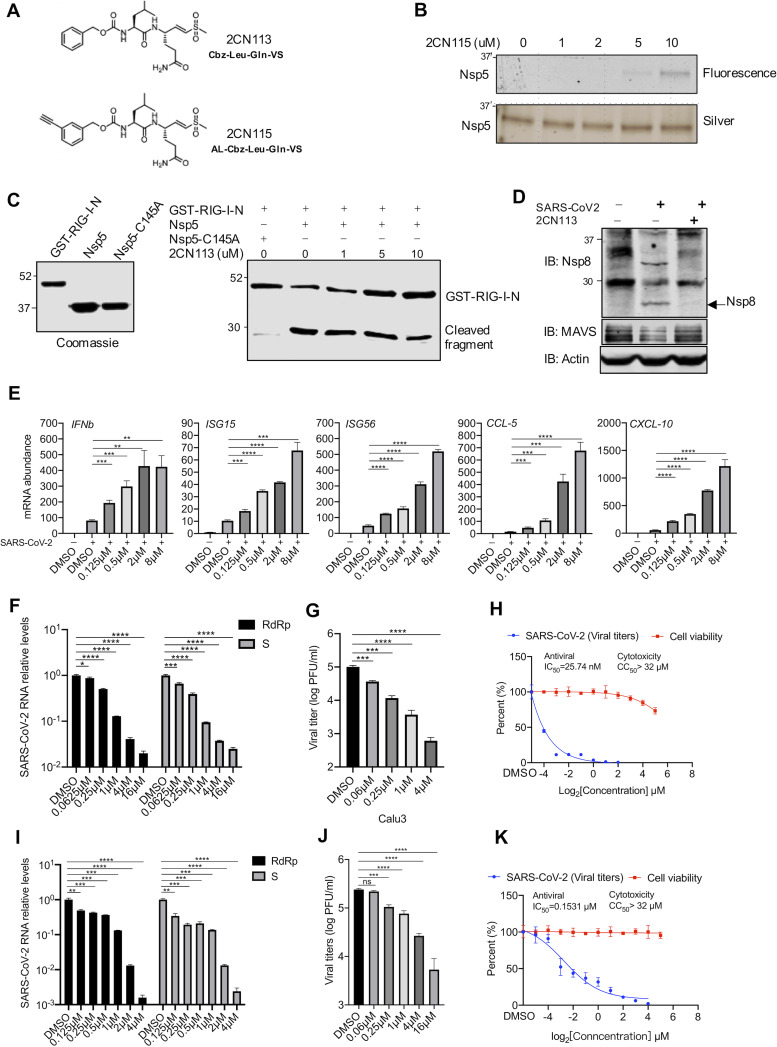
FIG 7.
A small-molecule inhibitor of Nsp5 restores innate immune activation and impedes SARS-CoV-2 replication. (A) Structures of 2CN113 and 2CN115. (B) Nsp5-Step-expressing 293T cells were treated with 2CN115 at the indicated doses for 2 h. Nsp5-Step was purified and subjected to binding analysis by in-gel fluorescence imaging and silver staining. (C) The effect of 2CN113 on in vitro GST–RIG-I-N cleavage by Nsp5 was analyzed with all proteins purified from E. coli by immunoblotting. (D) HeLa cells stably expressing hACE2 were treated with 2CN113 and infected with SARS-CoV-2 (MOI = 0.1). The effect of 2CN113 on SARS-CoV-2 Nsp8 processing was analyzed by immunoblotting with the indicated antibodies. (E) Calu-3 cells were treated with 2CN113 and infected with SARS-CoV-2 (MOI = 1). The mRNA abundance of antiviral genes was determined by real-time PCR at 72 h after SARS-CoV-2 infection. (F, G) Calu-3 cells were treated with 2CN113 and infected with SARS-CoV-2 (MOI = 0.1). The effect of 2CN113 on the SARS-CoV-2 RNA abundance (F) and viral titer (G) was determined at 72 h after SARS-CoV-2 infection by real-time PCR analysis of total RNA and plaque assay of the medium, respectively. S, SARS-CoV-2 S protein. (I, J) Vero cells stably expressing hACE2 were treated with 2CN113 and infected with SARS-CoV-2 (MOI = 0.001). The effect of 2CN113 on SARS-CoV-2 RNA abundance (I) and viral titer (J) was determined at 24 h after SARS-CoV-2 infection by real-time PCR analysis of total RNA and plaque assay of the medium, respectively. (H, K) The IC50 of 2CN113 on SARS-CoV-2 replication by plaque assay and the CC50 of 2CN113 on Calu-3 (H) and Vero (K) cells were determined and plotted. Data are means ± SD. Significance was calculated using Student’s two-tailed, unpaired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant. See also Fig. S6.