ABSTRACT
Future HIV-1 curative therapies require a thorough understanding of the distribution of genetically-intact HIV-1 within T-cell subsets during antiretroviral therapy (ART) and the cellular mechanisms that maintain this reservoir. Therefore, we sequenced near-full-length HIV-1 genomes and identified genetically-intact and genetically-defective genomes from resting naive, stem-cell memory, central memory, transitional memory, effector memory, and terminally-differentiated CD4+ T-cells with known cellular half-lives from 11 participants on ART. We find that a higher infection frequency with any HIV-1 genome was significantly associated with a shorter cellular half-life, such as transitional and effector memory cells. A similar enrichment of genetically-intact provirus was observed in these cells with relatively shorter half-lives. We found that effector memory and terminally-differentiated cells also had significantly higher levels of expansions of genetically-identical sequences, while only transitional and effector memory cells contained genetically-intact proviruses that were part of a cluster of identical sequences. Expansions of identical sequences were used to infer cellular proliferation from clonal expansion. Altogether, this indicates that specific cellular mechanisms such as short half-life and proliferative potential contribute to the persistence of genetically-intact HIV-1.
KEYWORDS: cell proliferation, cellular half-life, human immunodeficiency virus, persistence
INTRODUCTION
One of the major barriers to an HIV-1 cure is the persistence of replication-competent HIV-1 in memory CD4+ T-cells during antiretroviral therapy (ART) (1–3). Although ART reduces viremia to low levels, the virus persists in cellular reservoirs, and treatment cessation results in the resumption of viral replication (4, 5). Understanding the cellular mechanisms that contribute to the persistence of HIV-1 in cells during therapy will be vital to developing future curative strategies.
Previous studies have demonstrated that the number of HIV-1-infected cells remains remarkably stable during ART (3, 6), despite cell death and a lack of ongoing viral replication (7). Cellular proliferation has been found to contribute to the persistence of HIV-1 in memory T-cells (5, 6, 8–18), and this can occur through clonal expansion after exposure to cognate antigens, through binding of homeostatic cytokines, or as a result of the proviral integration site (19–22). However, these proliferative events are influenced by the diversity of the CD4+ T-cell compartment. T-cell subsets exist across a gradient of differentiation, from less differentiated naive (NV), stem-cell memory (SCM), and central memory (CM) populations to more differentiated transitional memory (TM), effector memory (EM), and terminally-differentiated (TD) cells (19, 23). This linear differentiation gradient reflects an increasing potential of T-cells to proliferate in response to cognate antigens.
Recent studies have shown that genetically-intact HIV-1 proviruses are unequally distributed across CD4+ T-cell subsets and anatomical compartments (17, 24, 25). We have previously observed higher levels of genetically-intact provirus in EM T-cells and cells expressing the activation marker human leukocyte antigen-antigen D related (HLA-DR) cells during long-term ART (24, 25). Analysis of the unique characteristics of each T-cell subset may inform how genetically-intact HIV-1 persists within these cells during ART. In particular, markers for CD4+ T-cell subsets at various maturation stages (26, 27) provide the opportunity to quantify the turnover of these discrete subsets and examine the relationship between cellular replacement rates and HIV-1 persistence during ART (28).
The timing of ART initiation may influence the level and genetic characteristics of persistent HIV-1 in treated individuals (6, 10). Early ART initiation limits the seeding of HIV-1 infection; however, studies have shown that this has little effect on HIV-1 disease outcome once ART is interrupted (29–33). How timing of ART initiation affects the level and distribution of genetically-intact proviruses in participants on effective ART is unclear and requires investigation.
In this study, we employed the full-length individual proviral sequencing (FLIPS) assay to identify genetically-intact HIV-1 proviral sequences within peripheral blood resting CD4+ T-cell subsets with known half-lives (24, 28). We found that T-cells with a relatively shorter half-life (i.e., EM and TM cells) have a higher proviral infection frequency. Importantly, this finding is associated with the turnover rate of a cell, suggesting that cellular replacement rates of specific cell subsets play a role in maintaining persistent HIV-1 across CD4+ T-cells. The half-life or replacement rate of a cell subset can include cells leaving or entering the compartment due to cell death, cellular differentiation, or migration into a different anatomical site. We will continue to refer to these collectively as cellular half-lives. Furthermore, we found that cells with shorter half-lives have higher levels of both genetically-identical sequences and genetically-intact identical sequence expansions, which suggests that cellular proliferation is an important mechanism for maintaining persistent HIV-1 in the peripheral blood during ART. We found no significant difference in the HIV-1 infection frequency and levels of genetically-intact HIV-1 provirus as a function of when ART was initiated. This study provides evidence that cellular mechanisms such as half-life and proliferation play important roles in maintaining genetically-intact HIV-1 during ART.
RESULTS
We obtained peripheral blood resting CD4+ T-cell subsets (NV, SCM, CM, TM, EM, and TD) from large blood draws in 11 participants, 6 who initiated ART <6 months after infection (early group) and 5 who initiated ART >6 months after infection (late group) (Table 1). The fractional replacement rates (that is, the proportion of cells replaced per day) of these cell subsets were estimated from in vivo deuterium labeling (28). This was used to calculate cellular half-lives for each cell subset in each participant (Table 2). The fractional replacement rates calculated for each cell subset was similar between 24 ART-suppressed HIV-1-infected participants and 6 uninfected participants, although the size of the cellular pool for each cell subset was lower in the ART-suppressed HIV-1-infected participants compared to uninfected participants (28). The FLIPS assay (24) was employed to sequence near-full-length HIV-1 genomes at the single-genome level. HIV-1 genomes were considered genetically-intact if they lacked genetic defects such as inversions, large internal deletions, hypermutation, deleterious stop codons, frameshifts or mutations in the packaging signal and/or major splice donor (MSD) site (cis-acting region) (Fig. 1A) (24).
TABLE 1.
Participant demographics
| Group and SCOPE PID | Sex | Age | Viral loada (copies/ml) | CD4+ T-cell counta (cells/μl) | Time of infection before initiation of therapy (mo) | ART durationa (yr) | ART durationa (classification) |
|---|---|---|---|---|---|---|---|
| Early | |||||||
| 2647 | Male | 33 | <40 | 532 | 4.5 | 3.4 | Short |
| 2531 | Male | 51 | <40 | 1,163 | 1.9 | 3.4 | Short |
| 2664 | Male | 46 | <40 | 637 | 4.1 | 2.7 | Short |
| 2606 | Male | 29 | <40 | 787 | 1.7 | 3.5 | Short |
| 2454 | Male | 35 | <40 | 513 | 0.7 | 7.1 | Long |
| 2661 | Male | 54 | <40 | 739 | 3.4 | 12.9 | Long |
| Late | |||||||
| 1408 | Male | 31 | <40 | 637 | 45.5 | 3.1 | Short |
| 3632 | Male | 31 | <40 | 902 | 20.8 | 1.8 | Short |
| 1756 | Male | 29 | <40 | 582 | 6.8 | 4.1 | Short |
| 2274 | Male | 54 | <40 | 486 | 13.1 | 11.8 | Long |
| 2208 | Male | 64 | <40 | 437 | 114.5 | 7.1 | Long |
At the time of sampling.
TABLE 2.
Cellular half-lives for each subset and participant
| Group and SCOPE PID | Half-life (days) |
|||||
|---|---|---|---|---|---|---|
| NV | SCM | CM | TM | EM | TD | |
| Early | ||||||
| 2647 | 488 | 160 | 103 | 94 | 88 | 165 |
| 2531 | 839 | 258 | 149 | 111 | 82 | 274 |
| 2664 | 2,484 | 184 | 202 | 143 | 151 | 536 |
| 2606 | 1,451 | 138 | 145 | 114 | 156 | 296 |
| 2454 | 2,146 | 139 | 135 | 105 | 108 | 245 |
| 2661 | 2,361 | 266 | 430 | 261 | 174 | 385 |
| Median | 1,799 | 172 | 147 | 113 | 130 | 285 |
| Late | ||||||
| 1408 | 1,747 | 153 | 144 | 95 | 64 | 291 |
| 3632 | 766 | 126 | 106 | 74 | 71 | 203 |
| 1756 | 1,107 | 165 | 93 | 77 | 52 | 244 |
| 2274 | 904 | 99 | 135 | 120 | 133 | 221 |
| 2208 | 557 | 89 | 128 | 97 | 98 | 238 |
| Median | 904 | 126 | 128 | 95 | 71 | 238 |
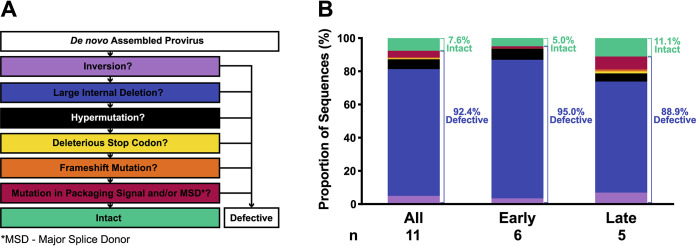
FIG 1.
Proportion of defective and intact viral DNA sequences isolated from each ART treatment group. (A) Sequence classification pipeline. Sequences were classified by a process of elimination that identified the major defects within each sequence. (B) Characteristics of sequences are colored the same as those for panel A. Number of participants is noted under each group.
Proviral characteristics.
A total of 1,277 viral sequences were isolated from the 11 participant samples. While the majority of these sequences (92.4%) were defective, we were able to identify 97 genetically-intact proviral sequences (7.6%) (Fig. 1B, Table 3). Of the 1,277 viral sequences, 727 were isolated for the early group, while 550 were isolated for the late group. In both groups, the majority of sequences were found to contain large internal deletions, comprising 95.0% of sequences in the early group and 88.9% of sequences in the late group. For the early and late groups, the proportion of hypermutated sequences was 6.7% and 4.9%, respectively (P = 0.79, mixed-effects logistic regression). We identified sequences in the late group that were defective due to a deleterious stop codon caused by a single-nucleotide polymorphism (1.3% of total sequences), and this type of defect was not found in the early group. We found 7.8% of the defective sequences from the late group contained a defect in the cis-acting region, whereas only 1.2% of these defects were found in the early group (P = 0.034, mixed-effects logistic regression). For the early and late groups, 5.0% and 11.1% of the total sequences were genetically-intact, respectively (P = 0.27, mixed-effects logistic regression).
TABLE 3.
Number of cells and sequences obtained for each subset and participant
| Group and SCOPE PID | No. of sequences and CD4+ T-cells used for analysis |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NV |
SCM |
CM |
TM |
EM |
TD |
|||||||||||||
| Cells analyzed | Defective | Intact | Cells analyzed | Defective | Intact | Cells analyzed | Defective | Intact | Cells analyzed | Defective | Intact | Cells analyzed | Defective | Intact | Cells analyzed | Defective | Intact | |
| Early | ||||||||||||||||||
| 2647 | 2,008,627 | 20 | 1 | 104,356 | 2 | 0 | 606,738 | 49 | 2 | 488,060 | 35 | 0 | 557,995 | 46 | 2 | 355,176 | 38 | 1 |
| 2531 | 846,802 | 2 | 1 | 505,614 | 11 | 1 | 845,044 | 26 | 0 | 369,349 | 33 | 1 | 241,380 | 37 | 0 | 589,001 | 33 | 2 |
| 2664 | 2,460,392 | 12 | 0 | NA | NA | NA | 1,124,804 | 42 | 0 | 750,758 | 24 | 1 | 285,373 | 56 | 0 | 350,372 | 39 | 0 |
| 2606 | 2,377,831 | NA | NA | NA | NA | NA | 3,074,632 | 10 | 0 | 2,709,673 | 34 | 1 | 1,419,446 | 21 | 0 | 794,481 | 2 | 0 |
| 2454 | 944,567 | 7 | 0 | NA | NA | NA | 822,853 | 34 | 0 | 606,953 | 42 | 1 | 370,087 | 26 | 0 | NA | NA | NA |
| 2661 | 1,312,753 | NA | NA | NA | NA | NA | 1,005,587 | 1 | 0 | 978,764 | 0 | 1 | 1,125,278 | 9 | 21 | NA | NA | NA |
| Late | ||||||||||||||||||
| 1408 | 2,008,675 | 1 | 0 | NA | NA | NA | 666,109 | 39 | 0 | 288,519 | 46 | 0 | 196,068 | 20 | 6 | 231,663 | 8 | 0 |
| 3632 | 1,972,938 | 6 | 2 | 155,591 | 3 | 0 | 844,189 | 14 | 0 | 1,200,376 | 31 | 2 | 409,945 | 35 | 0 | 677,269 | 3 | 0 |
| 1756 | 2,860,172 | NA | NA | 131,518 | 2 | 0 | 1,024,396 | 15 | 0 | 997,705 | 19 | 15 | 679,114 | 18 | 22 | 175,675 | NA | NA |
| 2274 | 342,384 | NA | NA | NA | NA | NA | 1,107,273 | 25 | 1 | 825,005 | 43 | 0 | 350,637 | 25 | 10 | NA | NA | NA |
| 2208 | 1,010,249 | 36 | 0 | NA | NA | NA | 367,777 | 35 | 1 | 419,586 | 31 | 2 | 267,978 | 34 | 0 | 248,955 | NA | NA |
HIV-1 is enriched in memory CD4+ T-cells with short half-lives.
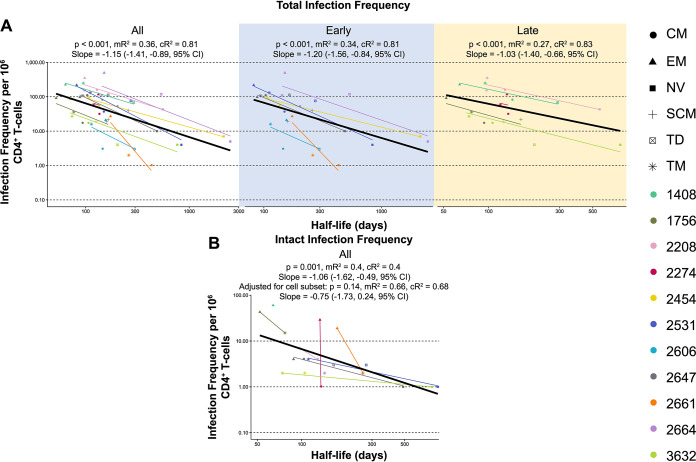
For each participant, we investigated whether the measured cellular half-lives of each CD4+ T-cell subset (Table 2) (28) were associated with HIV-1 infection frequencies. HIV-1 infection frequency was calculated for each participant and CD4+ T-cell subset (NV, SCM, CM, TM, EM, and TD) using the total number of sequences (intact or defective) amplified and the total number of cells collected for each subset. Cellular half-lives across all participants (Table 2) ranged from the longest in the NV subset (median, 1,107 days; interquartile range [IQR], 1,144 days) to the shortest in the EM subset (median, 98 days; IQR, 65.5 days). To determine the role of individual participant factors on infection frequencies, conditional and marginal R2 (cR2 and mR2, respectively) values were calculated for the association between cellular half-lives and HIV-1 infection frequencies. These indicate the amount of variability of infection frequency that can be accounted for by cellular half-life. The mR2 is the variability accounted for by the population-wide trend in half-life (Fig. 2, black lines). The cR2 is the variability accounted for by the trend in half-life after adjusting for individual participant effects (Fig. 2, individual participant colored lines).
FIG 2.
Infection frequency versus cellular half-life. (A) Mixed-effects modeling of total infection frequency per 106 cells versus cellular half-life (days) for all participants and early and late ART treatment groups. (B) Mixed-effects modeling of intact infection frequency per 106 cells versus cellular half-life (days) for all participants. The black line depicts simple regression analysis. P values and R2 values can be found for all participants and each ART treatment group. Two R2 values are calculated for this model, mR2 (marginal) and cR2 (conditional), which are similar to a linear regression comparing the relationship between half-life and infection frequency for individual participants or the population as a whole. Marginal R2 is calculated using all the data points on the graph regardless of participant, whereas the conditional R2 accounts for participant variability. The slopes and 95% CIs are reported for the population-wide trend.
Analysis of all participants demonstrated strong evidence for an association between cells with shorter half-lives and higher proviral loads (P < 0.001, slope = −1.15 [−1.41, −0.89, 95% confidence interval, or CI], mR2 = 0.36, cR2 = 0.81; Fig. 2A, mixed-effects linear regression). The trend between half-life and infection frequency is also present when the participants were divided into the separate early and late ART treatment groups (P < 0.001 for both, mR2 = 0.34 and cR2 = 0.81 for early, mR2 = 0.27 and cR2 = 0.83 for late; Fig. 2A, mixed-effects linear regression). When time to ART initiation (Table 1) was treated as a continuous variable, the trend between half-life and infection frequency remained after adjusting for time to ART initiation (data not shown).
For all participants, to assess whether the association between total infection frequency and cellular half-life is independent of other cellular subset effects, we next measured the association between HIV-1 infection frequency and cellular half-life while adjusting for cellular subset. We found that higher infection frequencies were still associated with shorter cellular half-lives (P = 0.026, slope = −0.86 [−1.63, −0.10, 95% CI], mR2 = 0.39, cR2 = 0.87). To investigate whether this relationship was driven by NV T-cells that exhibit both long half-lives and low infection frequencies, we repeated the analysis without these cells (see Table S1 in the supplemental material). This regression analysis without NV T-cells also revealed strong evidence that a cell containing an HIV-1 genome is more likely to have a shorter half-life (P < 0.001 without cellular subset adjustment, P = 0.043 with cellular subset adjustment). Together, this provides strong evidence that when accounting for participant variability, subsets with higher HIV-1 infection frequencies have shorter half-lives.
Linear regression analysis excluding NV T-cells. Download Table S1, DOCX file, 0.01 MB (13.4KB, docx) .
Copyright © 2021 Morcilla et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We next investigated the association between cellular half-life and the frequency of genetically-intact HIV-1 sequences. We found strong evidence that a cell containing a genetically-intact HIV-1 genome is more likely to have a shorter half-life (P = 0.001, slope = −1.06 [−1.62, −0.49, 95% CI], mR2 = 0.4, cR2 = 0.4) (Fig. 2B). This relationship exists in both the early and late groups separately (P = 0.016 and P = 0.032, respectively) (Fig. S1) and remains when NV T-cells were removed from the regression analysis (P = 0.018) (Table S1). We also investigated evidence for a relationship between genetically-intact infection frequency and cellular half-life after adjusting the data for cellular subset and found this trend was no longer statistically significant (P = 0.14, slope = −0.75 [−1.73, 0.24, 95% CI], mR2 = 0.66, cR2 = 0.68). We note that the low number of genetically-intact sequences isolated from each cellular subset (Table 3) means that this comparison has low statistical power.
Intact infection frequency versus cellular half-life for early and late groups. Mixed-effects modelling of intact infection frequency per 106 cells versus cellular half-life (days) for early and late ART treatment groups was used. The black line depicts simple regression analysis. P values and R2 values can be found for each analysis. Two R2 values are calculated for this model, cR2 (conditional) and mR2 (marginal), which are similar to a linear regression comparing the relationship between half-life and infection frequency for individual participants or the population as a whole. Marginal R2 is calculated using all the data points on the graph regardless of participant, whereas the conditional R2 accounts for participant variability. Slopes and 95% CIs are reported for the population-wide trends. Download FIG S1, TIF file, 1 MB (1MB, tif) .
Copyright © 2021 Morcilla et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expansions of identical sequences were found in more differentiated CD4+ T-cells.
Cellular proliferation has been shown to contribute to the persistence of HIV-1 proviruses in CD4+ T-cells (5, 6, 8–18). The presence of genetically-identical proviruses is likely to be indicative of cellular proliferation, as the error rate of the HIV-1 reverse transcriptase limits the transcription of identical proviruses by de novo replication (34). As such, when phylogenetic analyses were conducted for the HIV-1 sequences obtained from each participant sample, we inferred host cell proliferation through the presence of clusters consisting of ≥2 identical proviral sequences or expansions of identical sequences (EIS) (Fig. S2 and S3).
Phylogenetic tree of HIV-1 sequences from the early ART initiation group (participants 2647, 2531, 2664, 2606, 2661, and 2454). For each participant, a maximum likelihood phylogenetic tree is found in the center with subsets color coded: NV (blue), SCM (yellow), CM (green), TM (orange), EM (red), and TD (purple). In the inner concentric circle, sequences colored cyan are part of an EIS. The shading depicts the particular cell subset that forms the EIS, and grey shading reveals an EIS formed by multiple cell subsets. The outer concentric circle depicts full-length intact sequences by color corresponding to the cell subset containing that sequence. Sequence lengths are depicted in the outer ring. Download FIG S2, TIF file, 2.9 MB (3MB, tif) .
Copyright © 2021 Morcilla et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenetic tree of HIV-1 sequences from the late ART initiation group (participants 1408, 3632, 1756, 2274, and 2208). For each participant, a maximum likelihood phylogenetic tree is found in the center with subsets color coded: NV (blue), SCM (yellow), CM (green), TM (orange), EM (red), and TD (purple). In the inner concentric circle, sequences colored cyan are part of an EIS. The shading depicts the particular cell subset that forms the EIS, and grey shading reveals an EIS formed by multiple cell subsets. The outer concentric circle depicts full-length intact sequences by color corresponding to the cell subset containing that sequence. Sequence lengths are depicted in the outer ring. Download FIG S3, TIF file, 2.5 MB (2.6MB, tif) .
Copyright © 2021 Morcilla et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
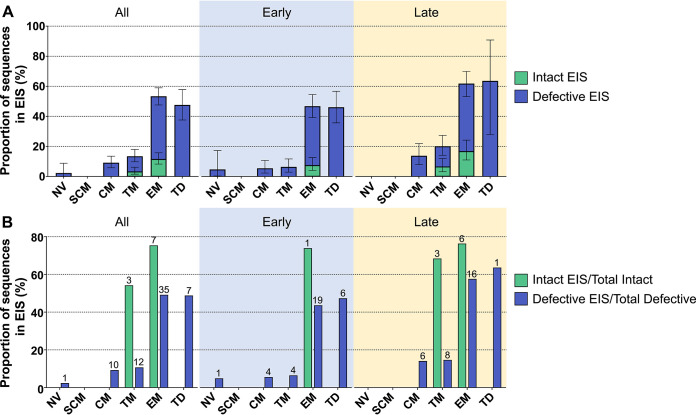
Sequences that were part of an EIS represented 27% of all sequences. The proportion of sequences belonging to an EIS was found to be highest in EM (53% of all EM sequences) and TD (48% of all TD sequences) compared to the other cell subsets studied (Fig. 3A) (P < 0.001, mixed-effects logistic regression). We note here that the proportion of sequences that were part of an EIS in the TD subset may have been reduced by potential contamination of this subset with NV T-cells. There was no difference in the overall proportion of sequences in an EIS between the early (24%) and late (31%) groups. However, when we compared the proportion of EIS between subsets, we observed TM to be significantly higher in the late group than the early group (P < 0.001, mixed-effects logistic regression). No other T-cell subset showed a statistically significant difference in the proportion of sequences belonging to an EIS between ART treatment groups (Table S2).
FIG 3.
Expansions of identical sequences for each ART treatment group. (A) Proportion of all sequences, either defective (blue) or intact (green), in an EIS in each CD4+ T-cell subset for all participants and early and late ART treatment groups. Data represented as means ± 95% CI. (B) The proportion of intact (green) and defective (blue) sequences that are also part of an EIS. The number of clusters that contribute to the pool of identical sequences is noted above the bars.
Comparisons between early and late ART treatment groups (mixed-effects logistic regression). Download Table S2, DOCX file, 0.01 MB (15.9KB, docx) .
Copyright © 2021 Morcilla et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genetically-intact sequences that were part of an EIS made up 4.6% of total sequences from the 11 participants. Only TM and EM cells contained genetically-intact sequences within an EIS. The proportion of genetically-intact sequences in an EIS was 3.6% and 12% of total sequences in TM and EM, respectively (Fig. 3A). There was no evidence for a difference in the overall proportion of genetically-intact sequences that were part of an EIS between the early and late groups (P = 0.23, mixed-effects logistic regression). Within each cell subset, there was no evidence for a difference in the number of genetically-intact sequences belonging to an EIS between the early and late groups (P > 0.2 for all, mixed-effects logistic regression) (Table S2).
We also assessed the contribution that sequences in an EIS have toward the total number of either genetically-intact or defective proviruses (Fig. 3B) (i.e., the number of sequences that were genetically-intact and part of an EIS over the total number of genetically-intact sequences and the number of sequences that were defective and part of an EIS over the total number of defective sequences). This was compared between cell subsets as well as between the early and late treatment groups. Although only 27% of all sequences were part of an EIS within the cells from the 11 participants, we found that 61% of all genetically-intact sequences belonged to an EIS. This finding suggests that genetically-intact sequences were disproportionately found within an EIS. Interestingly, 68% of the genetically-intact sequences in the TM subset were part of an EIS in the late group, while no genetically-intact sequences in an EIS were identified in the TM subset in the early group. However, this high proportion of genetically-intact identical sequences within the late group was formed by 3 clusters isolated from 1 participant, indicating that this result was driven by a single participant. The proportion of genetically-intact sequences in an EIS in EM was similar in the early (74%) and late groups (76%); however, this was also driven by a small number of clusters that contributed a large number of identical sequences. For EM, the proportion of defective sequences in an EIS was higher in the late group (58%) than the early group (44%) (P = 0.014, Fisher exact test). In the TD subset, we found the proportion of defective sequences in an EIS was 64% in the late group and 47% in the early group (P = 0.36, Fisher exact test). For the late group, this result came from one cluster of identical sequences from one participant.
Genetically-intact proviruses are unequally distributed in CD4+ T-cell subsets.
To determine whether genetically-intact proviruses would be concentrated in cell subsets with the shortest half-life (i.e., EM and TM), we assessed the frequency of HIV-1-infected cells (i.e., cells containing any HIV-1 genome, whether intact or defective) within resting NV, SCM, CM, TM, EM, and TD to determine the distribution of total and genetically-intact provirus within these T-cell subsets. Recent studies have investigated the distribution of genetically-intact HIV-1 provirus in NV, CM, TM, and EM subsets (24, 25), but the distribution of genetically-intact provirus in those that are resting as well as SCM and TD subsets has not been determined.
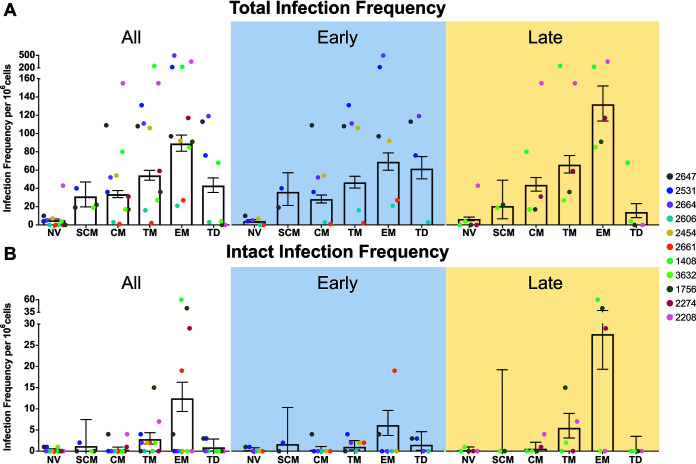
We first investigated the distribution of HIV-1-positive cells in each cell subset (Fig. 4A). We found strong evidence for a difference in the number of HIV-1-positive cells across the cell subsets (P < 0.001, mixed-effects logistic regression). We observed the order of this infection frequency to be EM>TM>TD/CM/SCM>NV (P < 0.005, mixed-effects logistic regression). There was a large range of infection frequencies measured between these subsets, with EM having an average of 89 proviruses per 106 cells and NV having an average of 5 proviruses per 106 cells (Fig. 4A). Within each cell subset, there was no evidence for a difference in estimated total infection frequency between the early and late groups (Table S2). When time to ART initiation (Table 1) was treated as a continuous variable, there was weak evidence for a positive association between total HIV-1 infection frequency and time to ART initiation in NV, CM, and EM, with the late treatment participants having a higher frequency of HIV-1 genomes in these cell subsets (0.05 < P = 0.10, mixed-effects logistic regression; Table S2).
FIG 4.
Infection frequency for each cell subset in each ART treatment group. (A) Total infection frequency per 106 cells for each CD4+ T-cell subset for all participants (n = 11) and each ART treatment group, early (n = 6) and late (n = 5). (B) Intact infection frequency per 106 cells for each CD4+ T-cell subset for all participants (n = 11) and each ART treatment group, early (n = 6) and late (n = 5). Data represented as mean ± 95% CI.
We investigated the distribution of intact HIV-1 provirus in each of the CD4+ T-cell subsets in the peripheral blood of the 11 participants (Fig. 4B). Interestingly, we were able to find intact provirus in all cell subsets studied, including SCM and TD. The total number of intact HIV-1 sequences ranged from 1 in SCM to 61 in EM (Table 3). Across all participants, there was no difference in intact infection frequency across cell subsets (P = 0.31, mixed-effects logistic regression), although EM was observed to have the highest intact infection frequency, followed by TM (Fig. 4B). Within each cell subset, there was no statistical evidence for a difference in the genetically-intact proviral infection frequency between the early and late participant groups (Table S2). This observation remained when time to ART initiation was treated as a continuous variable (Table S2).
DISCUSSION
Understanding how cellular mechanisms such as proliferation and half-lives contribute to the genetic composition of HIV-1 within infected cells is critical for the development of effective curative strategies. We therefore performed a detailed genetic analysis of HIV-1 proviral DNA sequences derived from a broad range of peripheral blood CD4+ T-cell subsets with measured cellular half-lives from participants who initiated ART during early or late infection. Moreover, the measurement of these cellular half-lives allowed us to assess the relationship between total and genetically-intact HIV-1 provirus and the cellular half-life of the CD4+ T-cell subsets in the peripheral blood (28). Taken together, these studies characterize the extent to which cellular half-lives and proliferation of CD4+ T-cell subpopulations contribute to the persistence of genetically-intact and defective HIV-1 provirus.
We found that HIV-1-infected cells are more likely to have a shorter half-life (i.e., TM or EM cells) than a longer half-life (i.e., NV and CM), and we saw a similar trend toward enrichment of cells containing genetically-intact provirus in cells with a shorter half-life. The in vivo labeling of T-cell subsets with deuterated glucose in uninfected volunteers revealed that there are distinct T-cell subpopulations with differing turnover rates, and EM T-cells were three times more proliferative than CM T-cells (35). The results of this study as well as our previous results (24) showed that EM and TM cells, which have shorter half-lives, have high levels of HIV-1 provirus, providing further evidence for the enrichment of HIV-1 provirus in cells with a shorter half-life. Our previous results also showed that EM and TM cells have high levels of genetically-intact provirus (24), and although we did not see a statistical difference in this study, we did see a trend toward high levels of genetically-intact provirus in TM and EM cells. It is important to note that few genetically-intact sequences were isolated from each cell subset in this study, making it difficult to observe a statistically significant relationship between the level of genetically-intact provirus and cellular subset or cellular half-life. Therefore, our findings indicate that the short half-lives and rapid turnover rates of memory T-cells, such as TM and EM, contribute to the level of total HIV-1 provirus they contain. This agrees with Bacchus-Souffan et al. (28), who found cells with shorter half-lives contain more HIV-1 DNA by measuring integrated HIV-1 DNA. Since measurements of total or integrated HIV-1 DNA measure mostly defective proviruses (24, 36), it is important to assess the relationship between cellular half-lives and genetically-intact proviruses, as this will inform the field how specific cellular mechanisms, such as half-lives/turnover rates, maintain this replication-competent reservoir in HIV-1-infected participants during effective ART. Interestingly, we also find that the short half-lives of memory T-cells may contribute to the level of genetically-intact provirus detected.
Cellular proliferation, as evidenced by expansions of genetically-identical HIV-1 sequences, has been shown by multiple studies to contribute to HIV-1 persistence during ART (5, 6, 8, 9, 11, 13–18, 24, 25, 37). In our study, we found that the proportion of sequences belonging to an expansion of genetically-identical HIV-1 sequences was highest in the more differentiated cells (EM and TD). In addition, the integration site sequencing analysis in the larger study of 24 participants (28) found that more differentiated cells had higher levels of clonally expanded integration sites. The proliferative capacity of EM has been described earlier (24, 25, 38); however, the proliferative capacity of TD is still not fully understood. One study found the proliferative capacity of TD cells to be low (39), while other studies found evidence for high proliferative capacity of TD cells by detecting T-cell receptor clonality and the expression of Ki67 (38, 40). Our data support the latter studies, as here we find relatively higher levels of genetically-identical sequences in TD compared with the other subsets.
In our study, we found that 61% of all genetically-intact sequences were part of an expansion of identical sequences, suggesting that cellular proliferation plays an important role in maintaining this genetically-intact proviral population. Despite the high level of genetically-identical sequences in EM and TD, we only found genetically-intact sequences that belonged to an expansion of identical sequences or EIS in the TM and EM subsets, with relatively higher levels found in the EM subset. This supports our previous findings that the EM subset has the highest level of genetically-intact sequences that were part of an EIS (24). The proliferative potential of memory T-cells increases exponentially as cells transition from CM to EM cells (28). In primates, TM cells have a lower magnitude of expansion than EM cells in response to interleukin-15 (IL‐15) in vivo (41, 42). Therefore, the potential for highly proliferative EM cells containing genetically-intact provirus to undergo clonal expansion may explain why we see higher levels of genetically-intact proviruses in cells with shorter half-lives and more rapid turnover rates and why fewer intact proviruses are part of an EIS in TM than in EM. This enrichment within proliferating cells with short half-lives is demonstrated by studies showing that HIV-1 can be found in memory CD4+ T-cell clones that tend to be activated more often, leading to the proliferation of these infected cells (22). These T-cell clones include those that recognize chronic/persistent viral antigens such as cytomegalovirus (CMV) or HIV-1 (43–45). It is, however, important to mention that these genetically-identical intact proviruses were found in certain participants but not others. Larger studies with more participants would provide more statistical power to further investigate the role that cellular turnover rate plays in the enrichment of genetically-intact proviruses within specific T-cell subsets. This cellular proliferation inferred from EIS could also be explained by insertional mutagenesis in oncogenes, resulting in the enhanced proliferation of some clones (16, 46). However, Bacchus-Souffan et al. (28) showed that only 1.3% of integration sites in the studied participants were found either in, or close to, oncogenes. Altogether, from our data we propose a model showing that shorter cellular half-life could be a predictor of a higher frequency of genetically-intact provirus in cells.
We found genetically-intact sequences in all subsets studied. Similar to our previous findings (24), we observed EM and TM cells to have the highest infection frequency with genetically-intact provirus, although this difference was not statistically significant in this study. This may be due to the fact that the 11 participants included in this study have been on ART for a shorter amount of time (median, 3.5 years; Table 1) than those included in the previous study (median, 15.8 years) (24). It may also be due to the small number of genetically-intact proviruses isolated from each cell subset in this study. There are currently no detailed studies looking at genetically-intact provirus for two of the T-cell subsets included in this study: the progenitor-like SCM (47) and the differentiated but CD45RA-expressing TD cells (39). While the relative contribution of proviruses from SCM and TD cells to the overall HIV-1 reservoir is low, our results reveal that these cells can contain genetically-intact genomes. This suggests that HIV-1 exploits both the progenitor-like properties of SCM (47) and the proliferative capacity of TD (38, 40) to persist during therapy. Collectively, all cell subsets studied can contribute to HIV-1 persistence; however, the high level of genetically-intact provirus in EM and TM suggests that cellular mechanisms specific to these two T-cell subsets, such as a short half-life, contribute to viral persistence.
It will be interesting to see if the association between the half-life of a cell and the presence of genetically-intact provirus will hold true for CD4+ T-cell subsets that were not included in this study. For example, we have previously shown that HLA-DR+ cells contain high levels of genetically-intact HIV-1 (25). This cell subset has been found to have a very short half-life in HIV-negative participants (28), which agrees with our finding that cell subsets with shorter half-lives contain high levels of genetically-intact proviruses. However, our study only included resting HLA-DR− CD4+ subpopulations, excluding events that occur during transient activation and HLA-DR expression of cells in vivo. Furthermore, a recent study has found clonally expanded, genetically-intact, and replication-competent provirus in functionally polarized Th1 cells in the peripheral blood, which were also not examined in our study (37). Lastly, all observations were restricted to circulating subpopulations of cells that are not representative of tissue-based cells, many of which do not recirculate. Less-characterized resident memory (TRM) T-cells are transcriptionally, phenotypically, and functionally distinct from recirculating memory cells and provide a first response against infections in tissue, where they accelerate pathogen clearance (48). The inclusion of HLA-DR+ cells, functionally polarized Th cells, and TRM cells in future analysis will help expand our knowledge of how cellular mechanisms affect HIV-1 persistence.
The 11 participants in our study were a subset of the 24 participants in a recently published study characterizing CD4+ T-cell turnover and HIV-1 persistence as measured by integrated HIV-1 DNA, cell-associated HIV-1 RNA, and proviral integration site sequencing (28). The study by Bacchus-Souffan et al. (28) found that shorter cellular half-lives were associated with higher total integrated DNA and cell-associated RNA levels, which supports our findings that cell subsets with shorter half-lives, such as TM and EM, have higher levels of HIV-1 infection and genetically-intact provirus. For the 11 participants included in our study, we did not observe differences in the levels of HIV-1-positive cells or cells containing genetically-intact HIV-1 between the early and late ART treatment groups. In contrast, the study of 24 participants found the late ART group had higher levels of both integrated HIV-1 DNA and cell-associated RNA in CM, TM, and EM T-cell subsets. The limited number of participants in our study (11 versus 24) may explain this discrepancy. It is also important to note that Bacchus-Souffan et al. (28) did not include TD in their analysis because the TD subset was contaminated with NV cells during cell sorting. We decided to include the TD results here since we found high levels of genetically-identical provirus in the TD subset, and this is not characteristic of NV cells (9, 24, 25). However, our results may represent a mixture of TD and NV T-cells. For future analyses, it would be important to conduct these analyses on a pure TD subset.
It may be expected that cell populations with short half-lives, which are more activated and turn over rapidly, would express more virus and, therefore, be cleared by the immune response. However, our study revealed that the level of genetically-intact HIV-1 is higher in cells with a short half-life such as TM and EM cells. One plausible explanation for this observation is that these cell populations with short half-lives are also more likely to express viral proteins, such as Nef. The expression of the Nef protein in cells during therapy has been shown by several studies to contribute to the persistence of HIV-1 by downregulating cell-surface major histocompatibility complex class I (MHC-I) and antigen presentation, which then allows the virus to evade CD8+ T-cell clearance (49, 50). In agreement with these findings, a recent study has revealed that HIV-1-specific T-cell response, and, more specifically, HIV-1 Nef-specific response continues during long-term ART. These HIV-1 Nef-specific responses were associated with the frequency of HIV-1-infected cells and indicate recent in vivo recognition of the HIV-1 Nef antigen during ART (51). Therefore, viral protein expression in more activated cells with a short half-life/rapid turnover rate could protect genetically-intact genomes, which retain the ability to express protective viral proteins. It is therefore possible that the expression of viral proteins such as Nef from genetically-intact proviruses is contributing to immune evasion and the persistence of these proviruses within cells with a short half-life such as TM and EM populations.
There are several limitations to our study that deserve comment. First, we did not employ in vitro studies to confirm the replication-competency of intact provirus identified by FLIPS. Therefore, we cannot be certain that the genetically-intact proviruses we identified are truly replication-competent. Second, it is important to note that NV T-cells, due to their long half-life, may affect the results of the regression analysis. However, regression analysis that did not include NV T-cells still showed a significant correlation between cell half-life and HIV-1 proviruses. Third, we assume that HIV-1-infected cells have fractional replacement rate characteristics similar to those of uninfected cells of the same maturational phenotype, which has not been confirmed. Finally, there is a lack of inclusion of female participants in this study, and it is known that sex plays an important role in modulating T-cell biology (52, 53).
In summary, the distribution of HIV-1 genomes across T-cell subsets during ART suggests that a short cellular half-life could be a predictor of a higher frequency of total HIV-1 proviruses or genetically-intact proviruses. Furthermore, cell subsets with high levels of genetically-intact provirus were marked by clusters of genetically-identical HIV-1 genomes, reflecting cellular proliferation. This indicates that specific cellular mechanisms such as a relatively short half-life and greater proliferative potential, characteristics of EM T-cells, contribute to the maintenance of genetically-intact HIV-1. A more complete understanding of these cellular mechanisms may inform future HIV-1 curative strategies.
MATERIALS AND METHODS
Participant cohort.
Eleven participants were sampled from a larger recently published study (28) of 24 ART-suppressed subtype B HIV-infected individuals who underwent deuterated water labeling and large-volume blood draws for evaluation of resting CD4+ T-cell turnover within sorted subsets. Participant characteristics are available in Table 1. Participants were divided into two groups based on early (<6 months) or late (>6 months) ART initiation.
In vivo labeling with deuterated (heavy) water.
Participants were provided with 70% deuterated water (2H20), with an oral intake of 50 ml three times daily for the first 7 days and 50 ml twice per day for the remainder of the 45-day-long labeling period as recently described (28). Peripheral blood was collected at baseline and at days 15, 30, and 45. Plasma was collected at all four time points, and saliva samples were collected at days 7 and 21 during the first month for assessment of body water enrichment, which was used to calculate the cellular replacement rate (28).
Cell sorting.
As previously described, fluorescence-activated cell sorting (FACSAria [BD Biosciences, Franklin Lakes, NJ, USA]) was used to sort resting (HLA-DR−) naive, stem-cell memory, central memory, transitional memory, effector memory, and terminally-differentiated CD4+ T-cell subsets (28). During the cell sorting, there was an estimated median of 20% contamination of the terminally-differentiated subset with naive cells based on an analysis of sjTREC content within each sorted population (28). Thus, the sorted terminally-differentiated population likely reflects a mix of naive and terminally-differentiated cells.
Measurement of cellular half-lives using deuterium enrichment.
The stable isotope/mass spectrometric method for measuring cell proliferation has been described previously (54). The cellular half-lives were calculated by determining deuterium enrichment in cellular genomic DNA as measured by gas chromatography mass spectroscopy (28). Controls required for working with low cell count samples were included. The inferred half-lives reflect the rate at which cells of each subpopulation in the peripheral blood turn over, through cell birth or death, cellular maturation into other phenotypes, or trafficking to or from anatomic compartments. The half-life for each cell subset from each participant can be found in Table 2.
FLIPS assay.
The FLIPS assay was performed as previously reported (24). Briefly, HIV-1 near-full-length proviruses (9 kb; 92% of the genome) were amplified at limiting dilution. Next-generation sequencing was conducted using the Illumina MiSeq platform. A median of 32 individual proviruses (range, 1 to 56) were sequenced per sorted CD4+ T-cell subset per participant (Table 3). Individual proviruses were assembled de novo using a specifically designed workflow in CLC Genomics (Qiagen). Proviruses were characterized as intact using a process of elimination that sequentially identified proviruses with inversions, large internal deletions (<8,900 bases), APOBEC3G-induced hypermutation, deleterious stop codons, frameshift mutations in any HIV-1 open reading frame (excluding tat exon 2 and nef) (55, 56), or a mutation in the packaging signal or MSD site (allowing for the presence of a cryptic MSD four nucleotides downstream of the MSD to salvage a mutated MSD [57]). Genomes were considered genetically-intact if they lacked such defects.
Further analysis of sequences.
EIS were identified using ElimDupes (Los Alamos database; https://www.hiv.lanl.gov/content/sequence/elimdupesv2/elimdupes.html). All sequences that were 100% identical were considered part of an EIS cluster. Maximum likelihood phylogenetic trees using the generalized time-reversible model were estimated for each participant using FastTree version 2.1.12 (58). Branch support was inferred using 1,000 bootstrap replicates, and gaps/deletions were weighted by proportion of nongaps. Annotated tree images were constructed using the iTOL software, version 5.5.1 (59).
Statistical analysis.
Statistical analysis was carried out in R version 3.6.3 (R Development Core Team, 2020). Mixed logistic regressions were carried out with the function glmer from the lme4 package (60). Mixed logistic regressions were used for testing of binary endpoints (e.g., EIS and infection frequencies) to account for correlated sequence information within participants. In these regressions, a random effect for the intercept was always included. Random effects for cellular subset were also included for infection frequency, intact infection frequency, and expansions of identical sequences models based on evidence for effect modification between participant and cellular subset (P < 0.001). The pairwise comparison of cellular subsets was tested using function glht from the multicomp package (61) on the mixed logistic regression, which automatically adjusts P values for multiple comparisons using the Bonferroni method. To compare the proportion of intact (or defective) sequences that were part of an EIS across early and late groups, the sequences from each participant were pooled and a Fisher exact test was used. Here, a mixed logistic regression to account for correlated data within an individual was not possible due to the limited number of intact sequences. The association between cellular half-life and proviral load was assessed with a linear mixed-effect model with function lmer in package lme4. The half-life values were log transformed before all analyses were performed. A random effect was included for intercept only. Conditional and marginal R2 (cR2 and mR2, respectively) values were calculated using function r.squaredGLMM in package MuMIn (62). These R2 values are analogous to R2 correlation values and indicate the amount of variability in infection frequency that can be accounted for by cellular half-life. mR2 is the variability accounted for by the population-wide trend in half-life (Fig. 2, black line). The cR2 is the variability accounted for by the trend in half-life after adjusting for individual participant effects (Fig. 2, individual participant colored lines).
Data availability.
The sequences analyzed in this study have been uploaded to GenBank under accession numbers MN466964 to MN467397, MZ922480 to MZ923010, and MZ962316.
ACKNOWLEDGMENTS
We thank the Sydney Informatics Hub (University of Sydney, Sydney, Australia) for support for and access to analytical software and the Australian Genome Research Facility (AGRF Sydney Site, the Westmead Institute for Medical Research, Sydney, Australia) for operating the MiSeq. We thank Yu Hsuan J. Dai, Andrea Wright, and Zoe Boyer for assisting with this study. We acknowledge with gratitude the participants who donated samples for this study.
This work was supported by the Delaney AIDS Research Enterprise (DARE) to Find a Cure (1U19AI096109 and 1UM1AI126611-01) and the Australian National Health and Medical Research Council (APP1061681 and APP1149990). The HOPE study was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01AI116368). The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We have no competing interests to declare.
Footnotes
Citation Morcilla V, Bacchus-Souffan C, Fisher K, Horsburgh BA, Hiener B, Wang XQ, Schlub TE, Fitch M, Hoh R, Hecht FM, Martin JN, Deeks SG, Hellerstein MK, McCune JM, Hunt PW, Palmer S. 2021. HIV-1 genomes are enriched in memory CD4+ T-cells with short half-lives. mBio 12:e02447-21. https://doi.org/10.1128/mBio.02447-21.
Contributor Information
Katie Fisher, Email: katie.fisher@sydney.edu.au.
Anne Moscona, Columbia University Medical College.
REFERENCES
- 1.Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 2.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 3.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF. 2003. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med 9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 4.Chun TW, Justement JS, Murray D, Hallahan CW, Maenza J, Collier AC, Sheth PM, Kaul R, Ostrowski M, Moir S, Kovacs C, Fauci AS. 2010. Rebound of plasma viremia following cessation of antiretroviral therapy despite profoundly low levels of HIV reservoir: implications for eradication. AIDS 24:2803–2808. doi: 10.1097/QAD.0b013e328340a239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Scheerder MA, Vrancken B, Dellicour S, Schlub T, Lee E, Shao W, Rutsaert S, Verhofstede C, Kerre T, Malfait T, Hemelsoet D, Coppens M, Dhondt A, De Looze D, Vermassen F, Lemey P, Palmer S, Vandekerckhove L. 2019. HIV rebound is predominantly fueled by genetically identical viral expansions from diverse reservoirs. Cell Host Microbe 26:347–358. doi: 10.1016/j.chom.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Stockenstrom S, Odevall L, Lee E, Sinclair E, Bacchetti P, Killian M, Epling L, Shao W, Hoh R, Ho T, Faria NR, Lemey P, Albert J, Hunt P, Loeb L, Pilcher C, Poole L, Hatano H, Somsouk M, Douek D, Boritz E, Deeks SG, Hecht FM, Palmer S. 2015. Longitudinal genetic characterization reveals that cell proliferation maintains a persistent HIV type 1 DNA pool during effective HIV therapy. J Infect Dis 212:596–607. doi: 10.1093/infdis/jiv092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bozzi G, Simonetti FR, Watters SA, Anderson EM, Gouzoulis M, Kearney MF, Rote P, Lange C, Shao W, Gorelick R, Fullmer B, Kumar S, Wank S, Hewitt S, Kleiner DE, Hattori J, Bale MJ, Hill S, Bell J, Rehm C, Grossman Z, Yarchoan R, Uldrick T, Maldarelli F. 2019. No evidence of ongoing HIV replication or compartmentalization in tissues during combination antiretroviral therapy: implications for HIV eradication. Sci Adv 5:eaav2045. doi: 10.1126/sciadv.aav2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee E, von Stockenstrom S, Morcilla V, Odevall L, Hiener B, Shao W, Hartogensis W, Bacchetti P, Milush J, Liegler T, Sinclair E, Hatano H, Hoh R, Somsouk M, Hunt P, Boritz E, Douek D, Fromentin R, Chomont N, Deeks SG, Hecht FM, Palmer S. 2020. Impact of antiretroviral therapy duration on HIV-1 infection of T cells within anatomic sites. J Virol 94:e01270-19. doi: 10.1128/JVI.01270-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel M-R, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy J-P, Haddad EK, Sékaly R-P. 2009. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Josefsson L, von Stockenstrom S, Faria NR, Sinclair E, Bacchetti P, Killian M, Epling L, Tan A, Ho T, Lemey P, Shao W, Hunt PW, Somsouk M, Wylie W, Douek DC, Loeb L, Custer J, Hoh R, Poole L, Deeks SG, Hecht F, Palmer S. 2013. The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc Natl Acad Sci USA 110:E4987–E4996. doi: 10.1073/pnas.1308313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bui JK, Sobolewski MD, Keele BF, Spindler J, Musick A, Wiegand A, Luke BT, Shao W, Hughes SH, Coffin JM, Kearney MF, Mellors JW. 2017. Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLoS Pathog 13:e1006283. doi: 10.1371/journal.ppat.1006283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simonetti FR, Sobolewski MD, Fyne E, Shao W, Spindler J, Hattori J, Anderson EM, Watters SA, Hill S, Wu X, Wells D, Su L, Luke BT, Halvas EK, Besson G, Penrose KJ, Yang Z, Kwan RW, Van Waes C, Uldrick T, Citrin DE, Kovacs J, Polis MA, Rehm CA, Gorelick R, Piatak M, Keele BF, Kearney MF, Coffin JM, Hughes SH, Mellors JW, Maldarelli F. 2016. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci USA 113:1883–1888. doi: 10.1073/pnas.1522675113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kearney MF, Wiegand A, Shao W, Coffin JM, Mellors JW, Lederman M, Gandhi RT, Keele BF, Li JZ. 2016. Origin of rebound plasma HIV includes cells with identical proviruses that are transcriptionally active before stopping of antiretroviral therapy. J Virol 90:1369–1376. doi: 10.1128/JVI.02139-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagner TA, McKernan JL, Tobin NH, Tapia KA, Mullins JI, Frenkel LM. 2013. An Increasing proportion of monotypic HIV-1 DNA sequences during antiretroviral treatment suggests proliferation of HIV-infected cells. J Virol 87:1770–1778. doi: 10.1128/JVI.01985-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohn LB, Silva IT, Oliveira TY, Rosales RA, Parrish EH, Learn GH, Hahn BH, Czartoski JL, McElrath MJ, Lehmann C, Klein F, Caskey M, Walker BD, Siliciano JD, Siliciano RF, Jankovic M, Nussenzweig MC. 2015. HIV-1 integration landscape during latent and active infection. Cell 160:420–432. doi: 10.1016/j.cell.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, Coffin JM, Hughes SH. 2014. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345:179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuo HH, Banga R, Lee GQ, Gao C, Cavassini M, Corpataux JM, Blackmer JE, Zur Wiesch S, Yu XG, Pantaleo G, Perreau M, Lichterfeld M. 2020. Blood and lymph node dissemination of clonal genome-intact human immunodeficiency virus 1 DNA sequences during suppressive antiretroviral therapy. J Infect Dis 222:655–660. doi: 10.1093/infdis/jiaa137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinzone MR, VanBelzen DJ, Weissman S, Bertuccio MP, Cannon L, Venanzi-Rullo E, Migueles S, Jones RB, Mota T, Joseph SB, Groen K, Pasternak AO, Hwang W-T, Sherman B, Vourekas A, Nunnari G, O'Doherty U. 2019. Longitudinal HIV sequencing reveals reservoir expression leading to decay which is obscured by clonal expansion. Nat Commun 10:728. doi:10.1038/s41467-019–08431-7. doi: 10.1038/s41467-019-08431-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanzavecchia A, Sallusto F. 2005. Understanding the generation and function of memory T cell subsets. Curr Opin Immunol 17:326–332. doi: 10.1016/j.coi.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 20.Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 21.Bruner KM, Cohn LB. 2019. HIV-1 reservoir dynamics in CD4+ T cells. Curr Opin HIV AIDS 14:108–114. doi: 10.1097/COH.0000000000000521. [DOI] [PubMed] [Google Scholar]
- 22.Liu R, Simonetti FR, Ho Y-C. 2020. The forces driving clonal expansion of the HIV-1 latent reservoir. Virol J 17:4. doi: 10.1186/s12985-019-1276-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sallusto F, Lanzavecchia A. 2011. Memory in disguise. Nat Med 17:1182–1183. doi: 10.1038/nm.2502. [DOI] [PubMed] [Google Scholar]
- 24.Hiener B, Horsburgh BA, Eden JS, Barton K, Schlub TE, Lee E, von Stockenstrom S, Odevall L, Milush JM, Liegler T, Sinclair E, Hoh R, Boritz EA, Douek D, Fromentin R, Chomont N, Deeks SG, Hecht FM, Palmer S. 2017. Identification of genetically intact HIV-1 proviruses in specific CD4(+) T cells from effectively treated participants. Cell Rep 21:813–822. doi: 10.1016/j.celrep.2017.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horsburgh BA, Lee E, Hiener B, Eden J-S, Schlub TE, von Stockenstrom S, Odevall L, Milush JM, Liegler T, Sinclair E, Hoh R, Boritz EA, Douek DC, Fromentin R, Chomont N, Deeks SG, Hecht FM, Palmer S. 2020. High levels of genetically intact HIV in HLA-DR+ memory T cells indicates their value for reservoir studies. AIDS 34:659–668. doi: 10.1097/QAD.0000000000002465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Appay V, van Lier RA, Sallusto F, Roederer M. 2008. Phenotype and function of human T lymphocyte subsets: consensus and issues. Cytometry A 73:975–983. doi: 10.1002/cyto.a.20643. [DOI] [PubMed] [Google Scholar]
- 27.Chattopadhyay PK, Roederer M. 2005. Immunophenotyping of T cell subpopulations in HIV disease. Curr Protoc Immunol Chapter 12:Unit 12.12. [DOI] [PubMed] [Google Scholar]
- 28.Bacchus-Souffan C, Fitch M, Symons J, Abdel-Mohsen M, Reeves DB, Hoh R, Stone M, Hiatt J, Kim P, Chopra A, Ahn H, York VA, Cameron DL, Hecht FM, Martin JN, Yukl SA, Mallal S, Cameron PU, Deeks SG, Schiffer JT, Lewin SR, Hellerstein MK, McCune JM, Hunt PW. 2021. Relationship between CD4 T cell turnover, cellular differentiation and HIV persistence during ART. PLoS Pathog 17:e1009214. doi: 10.1371/journal.ppat.1009214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ananworanich J, Schuetz A, Vandergeeten C, Sereti I, de Souza M, Rerknimitr R, Dewar R, Marovich M, van Griensven F, Sekaly R, Pinyakorn S, Phanuphak N, Trichavaroj R, Rutvisuttinunt W, Chomchey N, Paris R, Peel S, Valcour V, Maldarelli F, Chomont N, Michael N, Phanuphak P, Kim JH, Group RSS, on behalf of the RV254/SEARCH 010 Study Group . 2012. Impact of multi-targeted antiretroviral treatment on gut T cell depletion and HIV reservoir seeding during acute HIV infection. PLoS One 7:e33948. doi: 10.1371/journal.pone.0033948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Archin NM, Vaidya NK, Kuruc JD, Liberty AL, Wiegand A, Kearney MF, Cohen MS, Coffin JM, Bosch RJ, Gay CL, Eron JJ, Margolis DM, Perelson AS. 2012. Immediate antiviral therapy appears to restrict resting CD4+ cell HIV-1 infection without accelerating the decay of latent infection. Proc Natl Acad Sci USA 109:9523–9528. doi: 10.1073/pnas.1120248109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henrich TJ, Hatano H, Bacon O, Hogan LE, Rutishauser R, Hill A, Kearney MF, Anderson EM, Buchbinder SP, Cohen SE, Abdel-Mohsen M, Pohlmeyer CW, Fromentin R, Hoh R, Liu AY, McCune JM, Spindler J, Metcalf-Pate K, Hobbs KS, Thanh C, Gibson EA, Kuritzkes DR, Siliciano RF, Price RW, Richman DD, Chomont N, Siliciano JD, Mellors JW, Yukl SA, Blankson JN, Liegler T, Deeks SG. 2017. HIV-1 persistence following extremely early initiation of antiretroviral therapy (ART) during acute HIV-1 infection: an observational study. PLoS Med 14:e1002417. doi: 10.1371/journal.pmed.1002417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buzon MJ, Martin-Gayo E, Pereyra F, Ouyang Z, Sun H, Li JZ, Piovoso M, Shaw A, Dalmau J, Zangger N, Martinez-Picado J, Zurakowski R, Yu XG, Telenti A, Walker BD, Rosenberg ES, Lichterfeld M. 2014. Long-term antiretroviral treatment initiated at primary HIV-1 infection affects the size, composition, and decay kinetics of the reservoir of HIV-1-infected CD4 T cells. J Virol 88:10056–10065. doi: 10.1128/JVI.01046-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colby DJ, Trautmann L, Pinyakorn S, Leyre L, Pagliuzza A, Kroon E, Rolland M, Takata H, Buranapraditkun S, Intasan J, Chomchey N, Muir R, Haddad EK, Tovanabutra S, Ubolyam S, Bolton DL, Fullmer BA, Gorelick RJ, Fox L, Crowell TA, Trichavaroj R, O’Connell R, Chomont N, Kim JH, Michael NL, Robb ML, Phanuphak N, Ananworanich J, Phanuphak P, Teeratakulpisarn N, Chottanapund S, Fletcher J, Tantivitayakul P, Eamyoung P, Sutthichom D, Prueksakaew P, Puttamaswin S, Tipsuk S, Benjapornpong K, Ratnaratorn N, Munkong C, Tanjnareel K, Kanaprach R, Rerknimitr R, Wattanaboonyongcharoen P, Rojnuckarin P, Manasnayakorn S, Schuetz A, Akapirat S, RV411 Study Group, et al. 2018. Rapid HIV RNA rebound after antiretroviral treatment interruption in persons durably suppressed in Fiebig I acute HIV infection. Nat Med 24:923–926. doi: 10.1038/s41591-018-0026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Preston BD, Poiesz BJ, Loeb LA. 1988. Fidelity of HIV-1 reverse transcriptase. Science 242:1168–1171. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- 35.Macallan DC, Wallace D, Zhang Y, De Lara C, Worth AT, Ghattas H, Griffin GE, Beverley PCL, Tough DF. 2004. Rapid turnover of effector-memory CD4(+) T cells in healthy humans. J Exp Med 200:255–260. doi: 10.1084/jem.20040341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bruner KM, Murray AJ, Pollack RA, Soliman MG, Laskey SB, Capoferri AA, Lai J, Strain MC, Lada SM, Hoh R, Ho Y-C, Richman DD, Deeks SG, Siliciano JD, Siliciano RF. 2016. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med 22:1043–1049. doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee GQ, Orlova-Fink N, Einkauf K, Chowdhury FZ, Sun X, Harrington S, Kuo H-H, Hua S, Chen H-R, Ouyang Z, Reddy K, Dong K, Ndung'u T, Walker BD, Rosenberg ES, Yu XG, Lichterfeld M. 2017. Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. J Clin Invest 127:2689–2696. doi: 10.1172/JCI93289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corneau A, Cosma A, Even S, Katlama C, Le Grand R, Frachet V, Blanc C, Autran B. 2017. Comprehensive mass cytometry analysis of cell cycle, activation, and coinhibitory receptors expression in CD4 T cells from healthy and HIV-infected individuals. Cytometry B Clin Cytom 92:21–32. doi: 10.1002/cyto.b.21502. [DOI] [PubMed] [Google Scholar]
- 39.Oswald-Richter K, Grill SM, Leelawong M, Tseng M, Kalams SA, Hulgan T, Haas DW, Unutmaz D. 2007. Identification of a CCR5-expressing T cell subset that is resistant to R5-tropic HIV infection. PLoS Pathog 3:e58. doi: 10.1371/journal.ppat.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian Y, Babor M, Lane J, Schulten V, Patil VS, Seumois G, Rosales SL, Fu Z, Picarda G, Burel J, Zapardiel-Gonzalo J, Tennekoon RN, De Silva AD, Premawansa S, Premawansa G, Wijewickrama A, Greenbaum JA, Vijayanand P, Weiskopf D, Sette A, Peters B. 2017. Unique phenotypes and clonal expansions of human CD4 effector memory T cells re-expressing CD45RA. Nat Commun 8:1473. doi: 10.1038/s41467-017-01728-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lugli E, Goldman CK, Perera LP, Smedley J, Pung R, Yovandich JL, Creekmore SP, Waldmann TA, Roederer M. 2010. Transient and persistent effects of IL-15 on lymphocyte homeostasis in nonhuman primates. Blood 116:3238–3248. doi: 10.1182/blood-2010-03-275438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Picker LJ, Reed-Inderbitzin EF, Hagen SI, Edgar JB, Hansen SG, Legasse A, Planer S, Piatak M, Lifson JD, Maino VC, Axthelm MK, Villinger F. 2006. IL-15 induces CD4 effector memory T cell production and tissue emigration in nonhuman primates. J Clin Invest 116:1514–1524. doi: 10.1172/JCI27564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA. 2002. HIV preferentially infects HIV-specific CD4+ T cells. Nature 417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 44.Mendoza P, Jackson JR, Oliveira TY, Gaebler C, Ramos V, Caskey M, Jankovic M, Nussenzweig MC, Cohn LB. 2020. Antigen-responsive CD4+ T cell clones contribute to the HIV-1 latent reservoir. J Exp Med 217:e20200051. doi: 10.1084/jem.20200051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simonetti FR, Zhang H, Soroosh GP, Duan J, Rhodehouse K, Hill AL, Beg SA, McCormick K, Raymond HE, Nobles CL, Everett JK, Kwon KJ, White JA, Lai J, Margolick JB, Hoh R, Deeks SG, Bushman FD, Siliciano JD, Siliciano RF. 2021. Antigen-driven clonal selection shapes the persistence of HIV-1-infected CD4+ T cells in vivo. J Clin Invest 131:e145254. doi: 10.1172/JCI145254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner TA, McLaughlin S, Garg K, Cheung CY, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, Frenkel LM. 2014. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 345:570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buzon MJ, Sun H, Li C, Shaw A, Seiss K, Ouyang Z, Martin-Gayo E, Leng J, Henrich TJ, Li JZ, Pereyra F, Zurakowski R, Walker BD, Rosenberg ES, Yu XG, Lichterfeld M. 2014. HIV-1 persistence in CD4+ T cells with stem cell–like properties. Nat Med 20:139–142. doi: 10.1038/nm.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schenkel JM, Masopust D. 2014. Tissue-resident memory T cells. Immunity 41:886–897. doi: 10.1016/j.immuni.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blagoveshchenskaya AD, Thomas L, Feliciangeli SF, Hung CH, Thomas G. 2002. HIV-1 Nef downregulates MHC-I by a PACS-1- and PI3K-regulated ARF6 endocytic pathway. Cell 111:853–866. doi: 10.1016/s0092-8674(02)01162-5. [DOI] [PubMed] [Google Scholar]
- 50.Dirk BS, Pawlak EN, Johnson AL, Van Nynatten LR, Jacob RA, Heit B, Dikeakos JD. 2016. HIV-1 Nef sequesters MHC-I intracellularly by targeting early stages of endocytosis and recycling. Sci Rep 6:37021. doi: 10.1038/srep37021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stevenson EM, Ward AR, Truong R, Thomas AS, Huang SH, Dilling TR, Terry S, Bui JK, Mota TM, Danesh A, Lee GQ, Gramatica A, Khadka P, Alberto WDC, Gandhi RT, McMahon DK, Lalama CM, Bosch RJ, Macatangay B, Cyktor JC, Eron JJ, Mellors JW, Jones RB, AIDS Clinical Trials Group A5321 Team . 2021. HIV-specific T cell responses reflect substantive in vivo interactions with antigen despite long-term therapy. JCI Insight 6:e142640. doi: 10.1172/jci.insight.142640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meier A, Chang JJ, Chan ES, Pollard RB, Sidhu HK, Kulkarni S, Wen TF, Lindsay RJ, Orellana L, Mildvan D, Bazner S, Streeck H, Alter G, Lifson JD, Carrington M, Bosch RJ, Robbins GK, Altfeld M. 2009. Sex differences in the Toll-like receptor-mediated response of plasmacytoid dendritic cells to HIV-1. Nat Med 15:955–959. doi: 10.1038/nm.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scully EP, Gandhi M, Johnston R, Hoh R, Lockhart A, Dobrowolski C, Pagliuzza A, Milush JM, Baker CA, Girling V, Ellefson A, Gorelick R, Lifson J, Altfeld M, Alter G, Cedars M, Solomon A, Lewin SR, Karn J, Chomont N, Bacchetti P, Deeks SG. 2019. Sex-based differences in human immunodeficiency virus type 1 reservoir activity and residual immune activation. J Infect Dis 219:1084–1094. doi: 10.1093/infdis/jiy617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Busch R, Neese RA, Awada M, Hayes GM, Hellerstein MK. 2007. Measurement of cell proliferation by heavy water labeling. Nat Protoc 2:3045–3057. doi: 10.1038/nprot.2007.420. [DOI] [PubMed] [Google Scholar]
- 55.Foster JL, Garcia JV. 2007. Role of Nef in HIV-1 replication and pathogenesis. Adv Pharmacol 55:389–409. doi: 10.1016/S1054-3589(07)55011-8. [DOI] [PubMed] [Google Scholar]
- 56.Clark E, Nava B, Caputi M. 2017. Tat is a multifunctional viral protein that modulates cellular gene expression and functions. Oncotarget 8:27569–27581. doi: 10.18632/oncotarget.15174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Purcell DF, Martin MA. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol 67:6365–6378. doi: 10.1128/JVI.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bates D, Mächler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. J Stat Soft 67:48. doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- 61.Hothorn T, Bretz F, Westfall P. 2008. Simultaneous inference in general parametric models. Biom J 50:346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- 62.Bartoń K. 2020. MuMIn: multi-model inference. R package version 1.43.17. https://CRAN.R-project.org/package=MuMIn.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Linear regression analysis excluding NV T-cells. Download Table S1, DOCX file, 0.01 MB (13.4KB, docx) .
Copyright © 2021 Morcilla et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Intact infection frequency versus cellular half-life for early and late groups. Mixed-effects modelling of intact infection frequency per 106 cells versus cellular half-life (days) for early and late ART treatment groups was used. The black line depicts simple regression analysis. P values and R2 values can be found for each analysis. Two R2 values are calculated for this model, cR2 (conditional) and mR2 (marginal), which are similar to a linear regression comparing the relationship between half-life and infection frequency for individual participants or the population as a whole. Marginal R2 is calculated using all the data points on the graph regardless of participant, whereas the conditional R2 accounts for participant variability. Slopes and 95% CIs are reported for the population-wide trends. Download FIG S1, TIF file, 1 MB (1MB, tif) .
Copyright © 2021 Morcilla et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenetic tree of HIV-1 sequences from the early ART initiation group (participants 2647, 2531, 2664, 2606, 2661, and 2454). For each participant, a maximum likelihood phylogenetic tree is found in the center with subsets color coded: NV (blue), SCM (yellow), CM (green), TM (orange), EM (red), and TD (purple). In the inner concentric circle, sequences colored cyan are part of an EIS. The shading depicts the particular cell subset that forms the EIS, and grey shading reveals an EIS formed by multiple cell subsets. The outer concentric circle depicts full-length intact sequences by color corresponding to the cell subset containing that sequence. Sequence lengths are depicted in the outer ring. Download FIG S2, TIF file, 2.9 MB (3MB, tif) .
Copyright © 2021 Morcilla et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenetic tree of HIV-1 sequences from the late ART initiation group (participants 1408, 3632, 1756, 2274, and 2208). For each participant, a maximum likelihood phylogenetic tree is found in the center with subsets color coded: NV (blue), SCM (yellow), CM (green), TM (orange), EM (red), and TD (purple). In the inner concentric circle, sequences colored cyan are part of an EIS. The shading depicts the particular cell subset that forms the EIS, and grey shading reveals an EIS formed by multiple cell subsets. The outer concentric circle depicts full-length intact sequences by color corresponding to the cell subset containing that sequence. Sequence lengths are depicted in the outer ring. Download FIG S3, TIF file, 2.5 MB (2.6MB, tif) .
Copyright © 2021 Morcilla et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparisons between early and late ART treatment groups (mixed-effects logistic regression). Download Table S2, DOCX file, 0.01 MB (15.9KB, docx) .
Copyright © 2021 Morcilla et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The sequences analyzed in this study have been uploaded to GenBank under accession numbers MN466964 to MN467397, MZ922480 to MZ923010, and MZ962316.