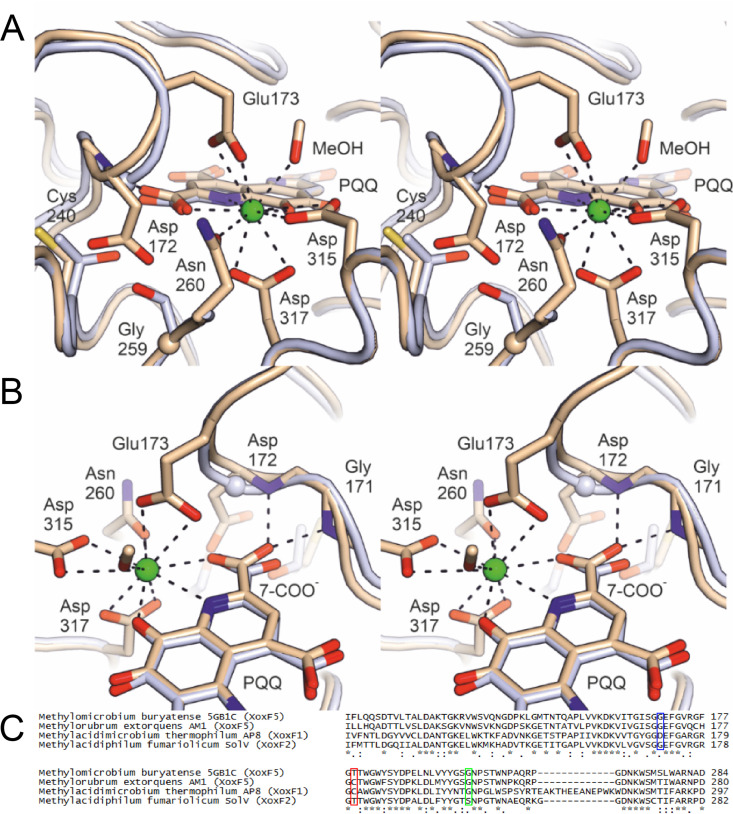
FIG 2.
(A) Stereofigures showing the active site of the B monomer in the crystal structure of the Nd-XoxF1 dimer (beige). Numbering of amino acid residues is according to the sequence of XoxF1 of M. thermophilum AP8. The structure of the cerium-containing XoxF2 of Methylacidiphilum fumariolicum SolV (light blue) is overlaid. Relevant residues and the PQQ molecule are shown as sticks, with glycine residues shown as spheres. The Nd atom is shown as a green sphere. The diagnostic Asp172 in Nd-XoxF1 is a glycine in the XoxF2 structure, as in all other MDHs of known structure. Opposite the Asp172 side chain, Nd-XoxF1 has a cysteine at position 240 and a glycine at position 259, which in the cerium-containing XoxF2 are a threonine and a serine, respectively. (B) As shown in panel A, but in a different orientation, providing a view of the loop containing the lanthanide-coordinating residue Glu173 (present in all known XoxF structures). The presence of the bulky Asp172 in this loop causes it to adopt a different conformation compared to XoxF2 of Methylacidiphilum fumariolicum SolV, affecting both Glu173 and the backbone amide N-H groups of Gly171 and Asp172, which engage in hydrogen bonds with the 7-carboxylate group of PQQ in Nd-XoxF1. This carboxylate, in turn, also coordinates the lanthanide. The alignment below compares the amino acid sequences near the active site of four XoxF-type MDHs of which the crystal structure was determined. Blue frame, Asp172 of M. thermophilum AP8 characteristic for XoxF1-type MDHs in contrast to glycine in the other structure; red frame, Cys240 of M. thermophilum AP8 to make space for the bulky Asp172 residue; green frame, Gly259 close to the active site which is a serine residue in the XoxF2 structure of Methylacidiphilum fumariolicum SolV. Asterisks indicate conserved amino acid residues; colons and dots indicate amino acid residues with highly and weakly similar properties, respectively.