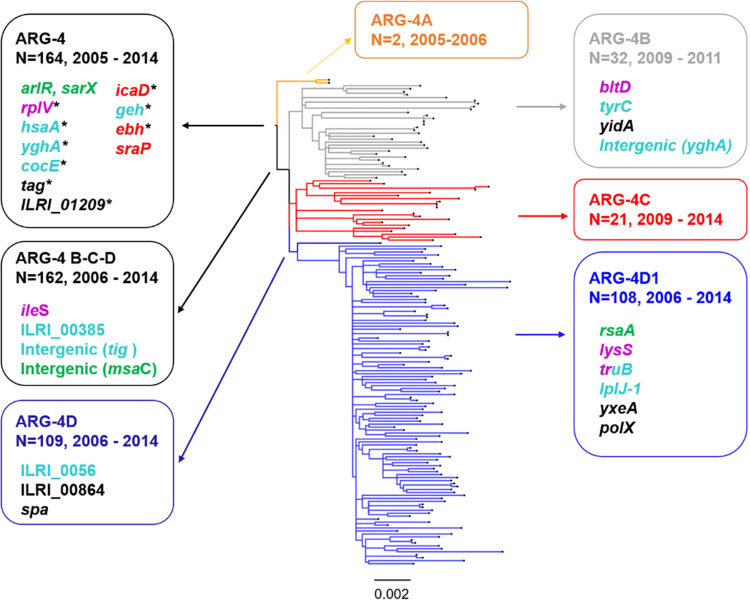
FIG 3.
Distribution of selected mutations in the phylogeny of clade ARG-4. Midpoint-rooted phylogenetic tree inferred as described for Fig. 1. Subclades within ARG-4 are colored in orange (ARG-4A), gray (ARG-4B), red (ARG-4C), and blue (ARG-4D). The scale bar represents the number of single nucleotide polymorphisms (SNPs) per variable site. The SNPs unique to each labeled subclade or internal node (arrows) are annotated within the boxes. Genes without an annotated name are in reference to the coding DNA sequence (CDS) of ILRI_Eymole1/1 reference genome by their locus tag. Gene names are colored by function as follows: protein synthesis (purple), response to free fatty acids and lipase geh (aqua), clumping/adhesion/biofilm (red), transcriptional regulators (green). SNPs leading to a premature stop codon are annotated with an asterisk.