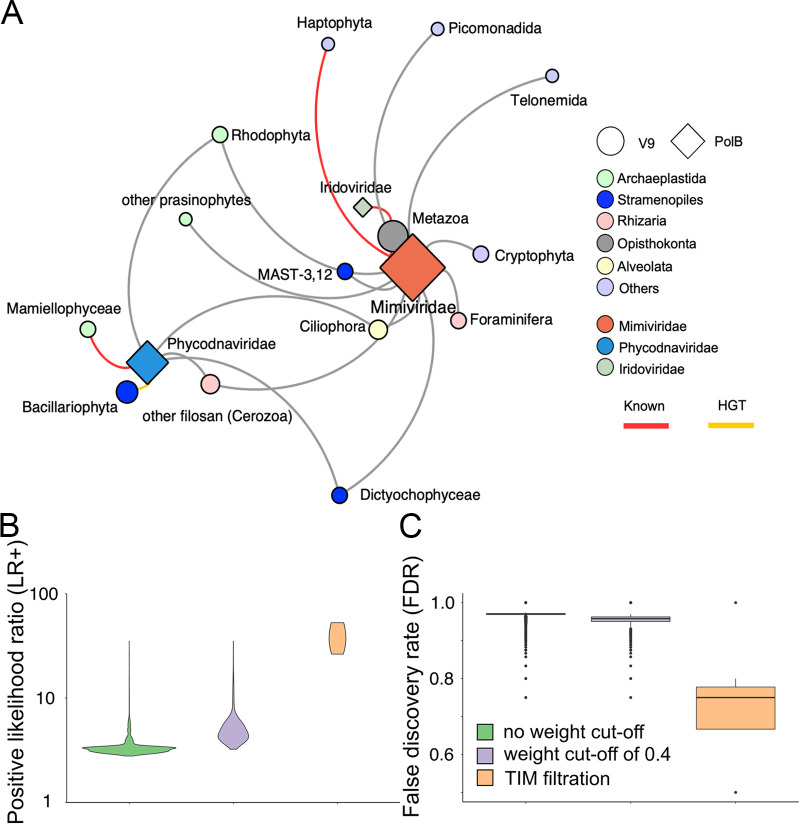
FIG 5.
Prediction of NCLDV virus-host relationships with TIM. (A) Undirected network that shows the relationships between NCLDVs and eukaryotes after TIM filtration. The size of each node indicates the number of predicted interactions of this group. The weight of network edges as defined by the number of tree nodes enriched in each viral family subtree to specific eukaryotic major lineages in the TIM analysis. Known virus-host relationships are highlighted in red, and the pairs found to have horizontal gene transfer are highlighted in yellow (1). (B) Performance of networks on NCLDV host prediction for original FlashWeave results without a weight cutoff, weight cutoff > 0.4, and TIM filtration, plotted by ggplot2 with a bandwidth of 2. (C) FDR of networks for NCLDV host prediction with the original FlashWeave results without a weight cutoff, weight cutoff > 0.4, and TIM filtration.