Fig. 5.
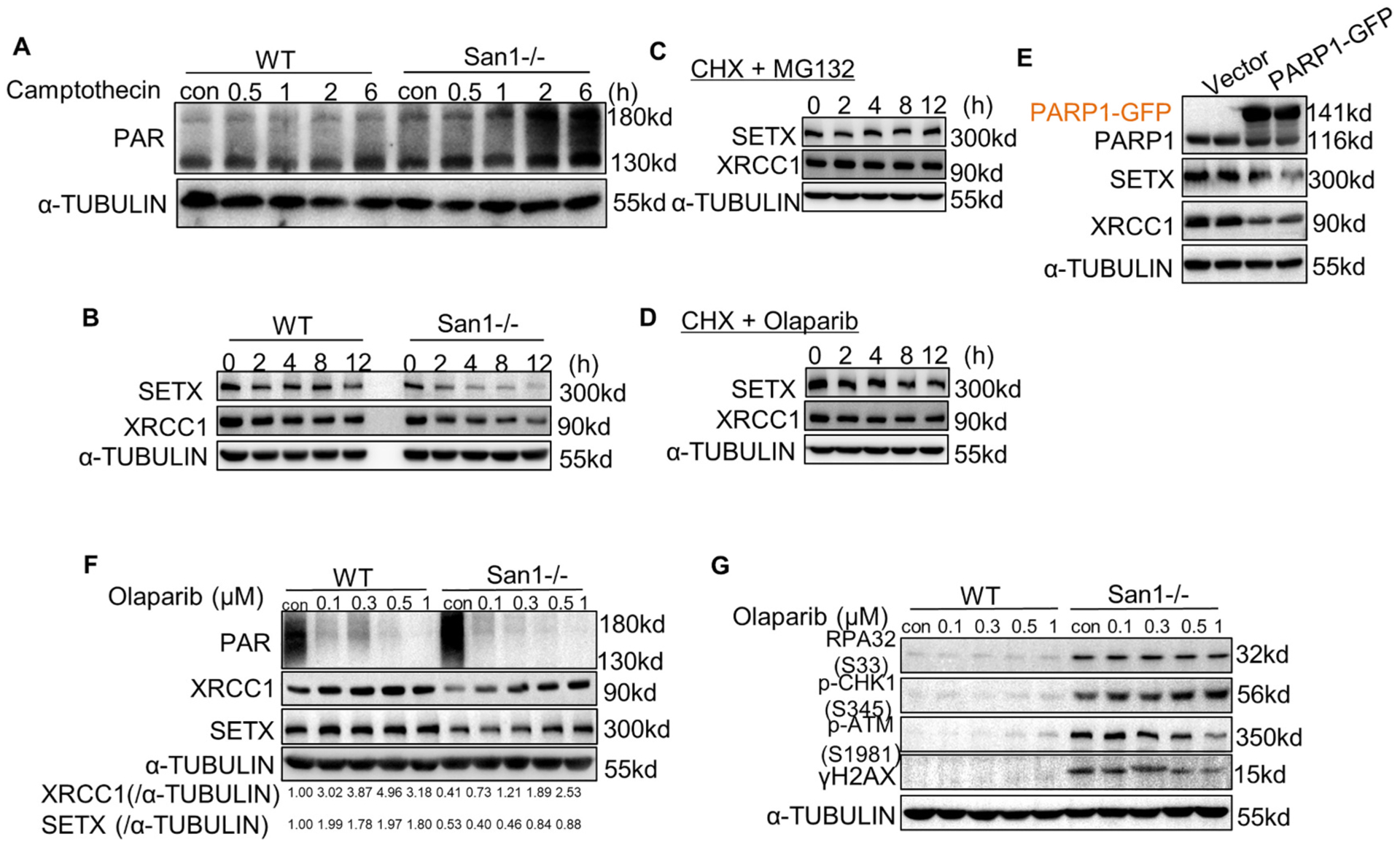
DDR forms a vicious circle with PARP1 overactivation, and Olaparib partially terminated this cycle. (A) San1−/− and WT cells were incubated with 1 μM camptothecin for indicated time. ADP-ribosylated proteins were determined by immunoblotting. (B) San1−/− and WT cells were incubated with 50 μM cycloheximide (CHX) for indicated time. SETX and XRCC1 were monitored by immunoblotting. (C) San1−/− cells were incubated with 50 μM cycloheximide (CHX) and 10 μM MG132 for indicated time. SETX and XRCC1 were detected by immunoblotting. (D) San1−/− cells were incubated with 50 μM cycloheximide (CHX) and 0.5 μM Olaparib for indicated time. SETX and XRCC1 were detected by immunoblotting. (E) WT cells were transfected with PARP1-GFP expression plasmid for 48 h. SETX and XRCC1 were detected by immunoblotting. (F and G) San1−/− and WT cells were incubated with Olaparib of different concentration gradients for 24 h. Indicators of DDR (p-ATM (S1981), γH2AX), replication stress (p-CHK1(S345), RPA32 (S33) and genomic processing proteins were monitored by immunoblotting.
