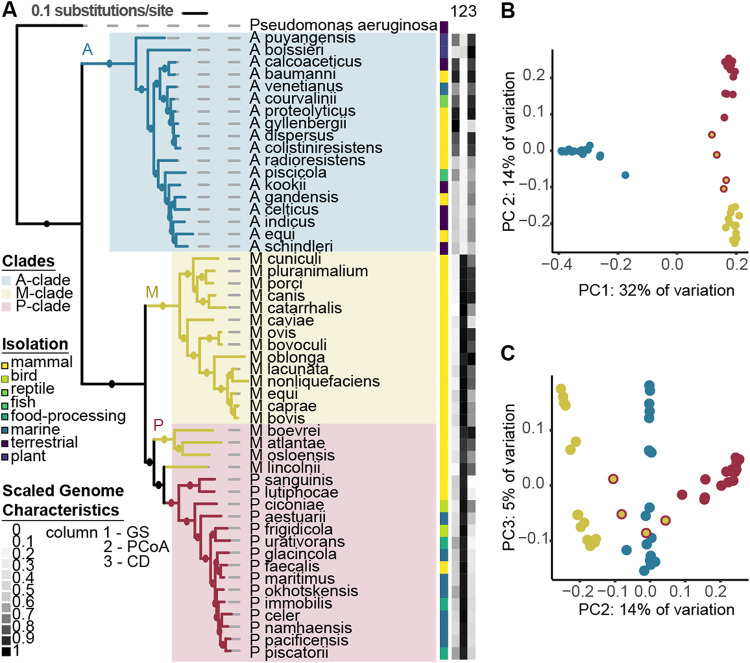
FIG 1.
Genomic and phenotypic diversity in the family Moraxellaceae. (A) The Moraxellaceae family phylogeny was constructed with 51 diverse Moraxellaceae genomes using the software PhyloPhlAn, which constructs a phylogeny using FastTree with 1,000 bootstraps, refined by RAxML under the PROTCATG model. Amino acid sequences from 400 marker genes were used in the alignment. Branches with bootstrap support greater than 70% are represented by filled circles. The scale bar represents the average amino acid substitutions per site. Pseudomonas aeruginosa was used as an outgroup. Clades are highlighted in colored blocks, and branches are colored by genus. Isolation source is depicted in a color strip, along with a heatmap of scaled notable genome characteristics that differ between the genera, with 0 representing the smallest value present and 1 the largest value (P. aeruginosa not included). GS, genome size, ranging between 1.8 Mb and 4.5 Mb. PCoA, PC1 values from a PCoA based on gene presence/absence data. CD, genome coding density, ranging from 80% to 89%. Growth temperature range data were collected from type strain publications. (B) PC1 and PC2 of a PCoA of a binary matrix of gene presence/absence for 51 species of Moraxellaceae, explaining 32% and 14% of the variation, respectively. Each genome is represented by one point, colored by genus. The P-clade Moraxella spp. are represented by yellow points with red outlines. (C) PC2 and PC3, explaining 14% and 5% of the variation, respectively.