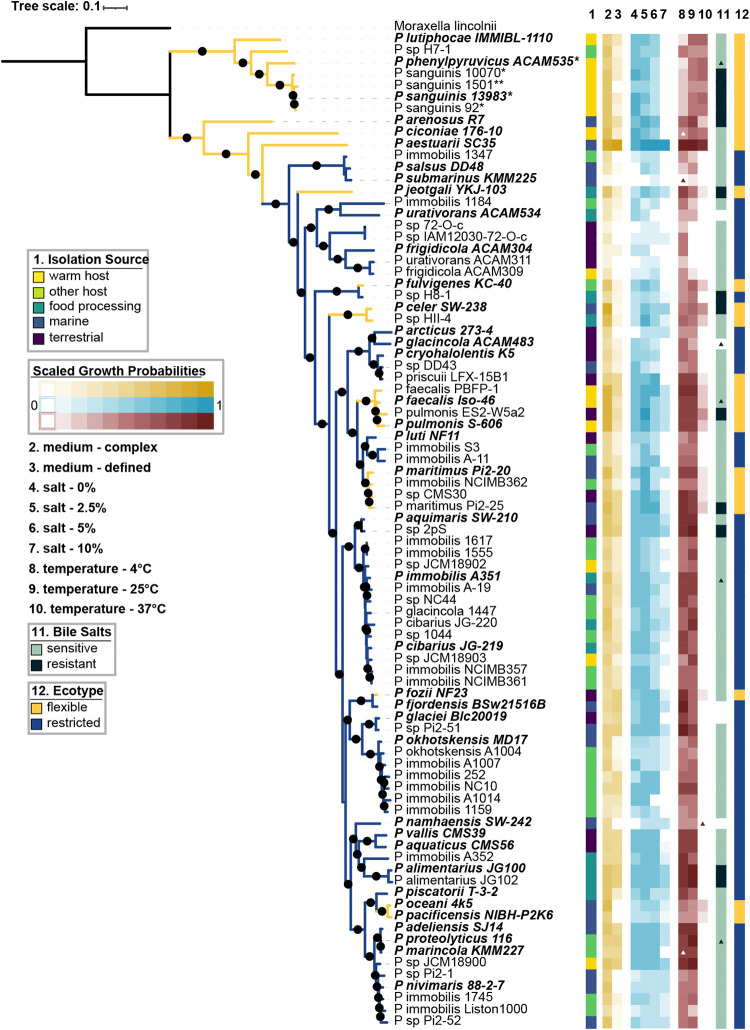
FIG 3.
Psychrobacter phenotypic and phylogenomic diversity. The Psychrobacter phylogeny was constructed using FastTree with 1,000 bootstraps, refined by RAxML under the PROTCATG model. Amino acid sequences from 400 marker genes were used in the alignment. Branches with bootstrap support greater than 70% are represented by filled circles. The scale bar represents the average amino acid substitutions per site. M. lincolnii was used as an outgroup. Type strain isolate names are indicated in bold and italicized type. Strains indicated with * next to their name exhibited growth defects in liquid media and were tested on solid agar media instead. Strains indicated with ** exhibited growth defects on solid and liquid media and were tested on solid media supplemented with 0.1% Tween 80. Isolation source is depicted in column 1 as a color strip. Columns 2 to 10 represent the growth probabilities of each strain for each condition; medium complexity is represented in yellow, salt concentration is represented in blue, and temperature is represented in red. Type strain data support our temperature data except where indicated—colored triangles show conditions under which we expected growth but did not observe it, while white triangles represent conditions under which we observed growth that we did not expect. Bile salt resistance is shown in column 11, with black triangles indicating strains where we expected resistance but observed sensitivity, and green triangles representing the opposite. The “ecotype” is shown in column 12 in a color strip and in the color of the branches.