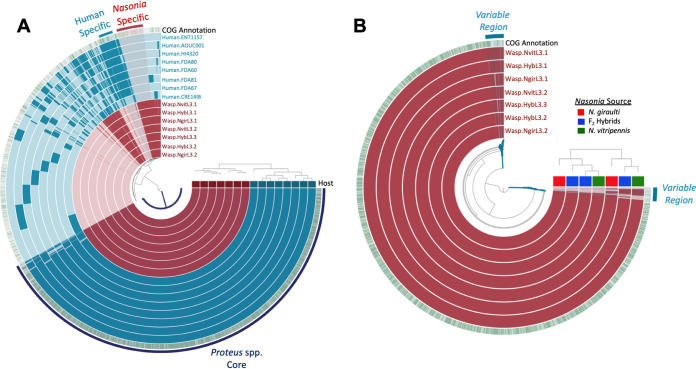
FIG 4.
Pangenome of Proteus mirabilis human and Nasonia isolates. (A) The 15 inner layers correspond to all 15 genomes, with Nasonia isolates shown in maroon and human isolates shown in blue. The top horizontal layer underneath the tree represents host source (Nasonia, maroon; human, blue). (B) The seven inner maroon layers correspond to the seven Nasonia-associated genomes sequenced as part of this study. The top horizontal layer underneath the tree represents host source (N. giraulti, red; F2 hybrids, blue; N. vitripennis, green). For both panels A and B, the solid bars in each of the inner circles show the presence of gene-clusters (GCs) in a given genome while the outer green circle depicts known COGs (green) versus unknown (white) assignments. The outermost layer of solid bars and text highlight groups of GCs that correspond to the genome core or to group-specific GCs. Genomes are ordered based on their gene-cluster distribution across genomes, which is shown at the top right corner (tree). Central dendrograms depict protein cluster hierarchy when displayed as protein cluster frequency.