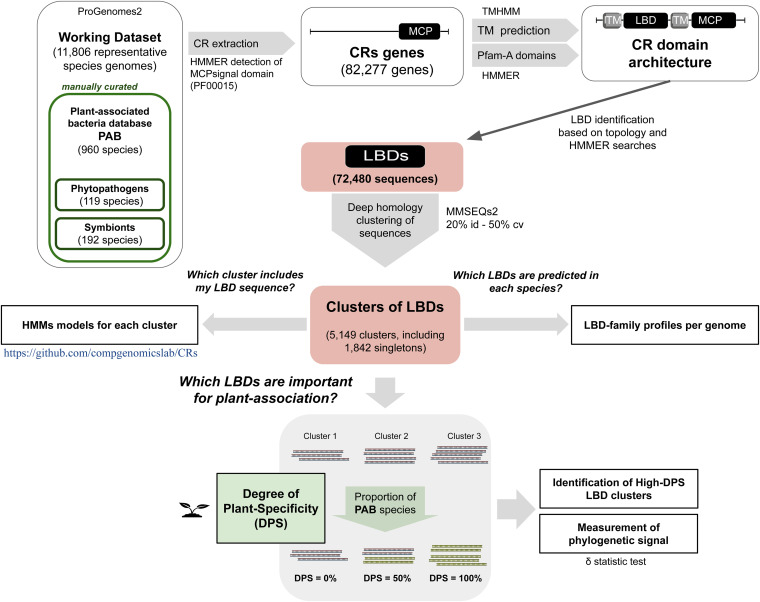
FIG 1.
Schematic view of the bioinformatics pipeline used to identify CRs that are potentially relevant for plant association. From a set of 11,806 representative prokaryotic genomes, 82,277 protein sequences were mined using HMM-based searches against the MCPsignal Pfam domain (PF00015). CR topology was analyzed by predicting transmembrane regions (TMs) and Pfam domains. Based on the topological analysis, LBD regions were predicted and a set of 72,480 LBD sequences was obtained. Clustering of LBDs based on sequence homology (20% minimum sequence identity with at least 50% sequence coverage) resulted in 5,149 clusters or subfamilies of LBDs, of which 1,842 contained a single sequence. To study a possible link between the LBD profiles and plant-associated lifestyle, a manually curated subset of 960 representative species of plant-associated bacteria (PAB) was generated, including phytopathogen (119) and symbiont (192) subsets. The determination of the proportion of PAB LBDs present in each cluster allowed us to assign the degree of plant specificity (DPS) value for each LBD subfamily. Subsequent analysis of high-DPS clusters identified LBD clusters that are potentially important for bacterium-plant associations. Furthermore, the validation of the high-DPS clusters as good ecological indicators was corroborated by measuring their phylogenetic signal. A detailed step-by-step description of the process can be found in Materials and Methods.