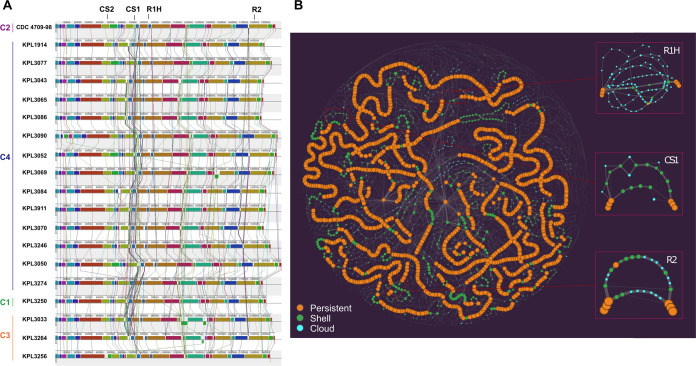
FIG 2.
D. pigrum displays conserved chromosomal synteny. (A) A MAUVE alignment of 19 closed D. pigrum genomes, with representatives from the four major clades in Fig. 1, shows a conserved order of chromosomal blocks across the phylogeny of strains collected 20 years apart. Vertical bars represent clades: clade 1, green; clade 2, purple; clade 3, orange; and clade 4, blue. CS1 and CS2 designate the CRISPR-Cas sites (Fig. 8), R1H represents the RM system insertional hot spot and R2 represents the site containing either a type II m5C RM system or a type IV restriction system (Fig. 7). (B) This PPanGGOLiN partitioned pangenome graph displays the overall genomic diversity of the 28 D. pigrum genomes. Each graph node corresponds to a GC; the node size is proportional to the total number of genes in a given cluster, and the node color represents the PPanGGOLiN assignment of GCs to the partitions: persistent (orange), shell (green), and cloud (blue). Edges connect nodes that are adjacent in the genomic context and their thickness is proportional to the number of genomes sharing that neighboring connection. The insets on the right depict subgraphs for sites R1H, CS1, and R2 showing several branches corresponding to multiple alternative shell and cloud paths. These sites with higher genomic diversity are surrounded by longer regions with conserved synteny, i.e., long stretches of consecutive persistent nodes (GCs). The static image depicted here was created with the Gephi software (https://gephi.org) (133) using the ForceAtlas2 algorithm (134) with the following parameters: scaling = 20,000, stronger gravity = true, gravity = 6.0, LinLog mode = true, and edge weight influence = 2.0.