ABSTRACT
Understanding mechanisms governing the resistance and resilience of microbial communities is essential for predicting their ecological responses to environmental disturbances. Although we have a good understanding of such issues for soil and lake ecosystems, how ecological resistance and resilience regulate the microbiota in the fish gut ecosystem remains unclear. Using the zebrafish model, we clarified the potential mechanisms governing the gut microbiota after exposure to silver nanoparticles (AgNPs). Here, we explored the ecological resistance and resilience of gut microbiota in zebrafish exposed to different concentrations of AgNPs (i.e., 10, 33 and 100 μg/liter) for 15, 45, 75 days. The high-throughput sequencing analysis of the 16S rRNA gene showed that AgNP exposure significantly reduced the α-diversity of gut microbiota and resulted in obvious dynamics of community composition and structure. However, the rebound of zebrafish gut microbiota was pushed toward an alternative state after 15 days of AgNP exposure. We found that homogeneous selection was a more prevalent contributor in driving gut community recovery after AgNP exposure. The resilience and resistance of gut microbiota responses to AgNP disturbance might be mainly determined by the predominant keystone taxa such as Acinetobacter and Gemmata. This study not only expanded our understanding of fish gut microbiota’s responses to pollutants but also provided new insights into maintaining host-microbiome stability during environmental perturbations.
IMPORTANCE Understanding the ecological mechanisms governing the resistance and resilience of microbial communities is a key issue to predict their responses to environmental disturbances. Using the zebrafish model, we wanted to clarify the potential mechanisms governing the resistance and resilience of gut microbiota after exposure to silver nanoparticles (AgNPs). We found that AgNP contamination significantly reduced the α-diversity of gut microbiota and resulted in obvious changes in community composition. The resilience and resistance of gut microbiota to AgNPs might be associated with the predominant keystone taxa (e.g., Acinetobacter and Gemmata). This study greatly expanded our understanding of how fish gut microbiota responds to environmental perturbations and maintains stability.
KEYWORDS: zebrafish, gut microbiota, resistance, resilience, silver nanoparticles
INTRODUCTION
The ecological stability of the microbial community as reflected by resistance and resilience to environmental changes or disturbances is a key issue for understanding ecosystem functions (1, 2). Microorganisms not only are the crucial players in biogeochemical cycling in different ecosystems but are also found to be linked with impairment of ecosystem functions when some microbial taxa are lost (3). Recent studies suggested that microbial communities could also be used for predicting ecosystem responses to global changes (4, 5). However, we still lack a full understanding of how microbial communities respond to ecological disturbances in particular ecosystems or global process models. Therefore, the resistance and resilience of microbial communities during environmental disturbances have received increasing interest during the last 2 decades.
Although the dynamics of taxonomic composition and community diversity are commonly involved in studying microorganisms’ responses to various disturbances, the microbial community assembly processes are also major matters for microbial resistance and resilience (6). Unfortunately, the underlying mechanisms of microbial community assembly and succession subjected to environmental disturbances still remain controversial (1). Generally, ecological patterns of communities can be visualized in four types of processes: selection, drift, speciation, and dispersal (7). It is commonly accepted that selection is a major force shaping microbial communities, which may help microorganisms withstand or adapt to a disturbance (8, 9). Microorganisms are also well-known materials for rapid dispersal in fluid ecosystems (10), and the dispersed members represent an important regional species pool that can colonize in disturbed ecosystems (3). Microbial dispersal can also enhance compositional recovery by reintroducing species that were lost after disturbances. For example, dispersal promoted microbial recovery before a drought episode by reintroducing sensitive taxa at the early stage of rewetting (11). However, the ecological processes governing microbial communities are still not easy to quantify accurately due to some methodological issues (12).
Currently, the potential mechanisms affecting microbial interactions are also used to address the microbial responses to environmental disturbances (13–15). Network analysis has become one of the most common approaches in exploring microbial cooccurrence patterns (16). A disturbance may lead to an increase in the abundance of some taxa as a consequence of the loss of major competitors that could not withstand the disturbance. Moreover, the modularity, average clustering coefficient (avgCC), average path distance (GD), and node centrality of networks are also good properties for characterizing microbial interactions and reflecting microbial resilience (3, 17). For example, a network-based analysis of bacterioplankton communities showed that networks with higher modularity would be more stable than dispersal networks (18). Similarly, the indirect effects of drought could result in lower stability of bacterial networks due to an increase in node centrality but a decrease in modularity (19–21). Thus, network analysis of microbial interactions can also expand our understanding of resistance and resilience during disturbances.
Recently, the extensive use of nanoparticles has resulted in environmental disturbances due to their toxic responses to biological systems (22). Nanoparticles may come from many consumer products, including textiles, cosmetics, plastic, food packaging, and medical appliances (23). Such products are potential sources of environmental release of nanoparticles into aquatic environments, which could pose risks to both natural ecosystems and organisms therein (24). Recent studies on zebrafish (Danio rerio) and common carp (Cyprinus carpio) showed that silver nanoparticles (AgNPs) were ingested and highly accumulated in the intestinal tract to cause gut microbial dysbiosis during acute toxic exposure (25, 26). However, whether gut microbiota affected by AgNP exposure could return to the initial state after environmental recovery is unclear. This study aimed to reveal the ecological resistance and resilience of gut microbiota in fish subjected to disturbance by the AgNPs.
The zebrafish has been an attractive vertebrate model for studying gut microbiota due to its small size, rapid development, optimum breeding, and maintenance conditions (27, 28). It has greatly increased our understanding of the mechanisms governing fish gut microbiota in the past 2 decades (for examples, see reference 29 to 31), which also can be used to investigate the responses of microbial communities to environmental stresses (32). Moreover, the gut ecosystem represents an isolated oceanic “island,” where the microbial community colonizes in the “ocean” of the surrounding environment (33, 34). This study, therefore, used the zebrafish model to clarify the mechanisms of microbial resistance and resilience. We hypothesize that the resistance and resilience of gut microbiota are mainly governed by selecting adaptive taxa and increasing their interactions in response to AgNP disturbances. To test this hypothesis, we used exposures to different concentrations of AgNPs (i.e., 10, 33, and 100 μg/liter) as disturbances to explore their possible effects and the underlying mechanisms maintaining gut microbiota. This study not only increases our understanding of how environmental disturbances affect the fish gut microbiota but also provides novel insights into maintaining stable gut microbiota for host health.
RESULTS
Gut microbial diversity and structure in relation to resistance and resilience.
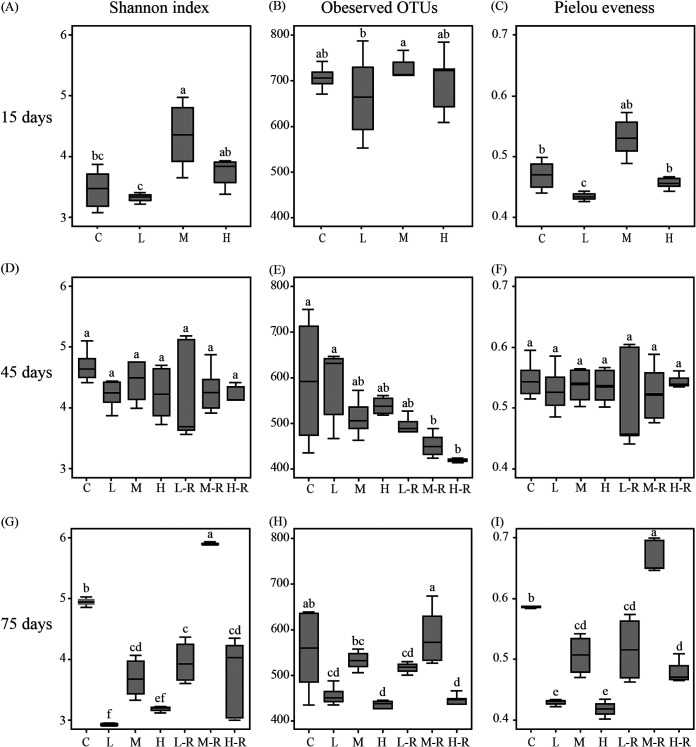
Through sequencing efforts with 14,285 sequences per sample, we classified a total of 6,927 operational taxonomy units (OTUs) (UPARSE, 97% cutoff) for the 105 samples. Zebrafish exposure to AgNPs for 15 days resulted in significant variations in the gut microbiota (Fig. 1). Specifically, the medium concentration exposure (M; 33 μg/liter) showed the highest α-diversity (Fig. 2A to C), while the low concentration exposure (L; 10 μg/liter) showed the lowest (P < 0.05) after AgNP disturbance. Also, there were significant differences in the Shannon and Pielou evenness indexes between the L and high exposure concentration (H; 100 μg/liter) groups (P < 0.05). At 45 days, the α-diversity of exposure groups was similar to that of the controls (Fig. 2D to F). In contrast, the Shannon index, observed OTUs, and Pielou evenness index of recovery of groups with low (L-R), medium (M-R), and high (H-R) concentrations of exposure were increasing, as measured at 75 days (Fig. 2G to I). The α-diversity of the M-R group was significantly higher (P < 0.05) than that of the continued-exposure groups and controls. So, AgNP disturbance significantly decreased the Shannon index, observed OTUs, and Pielou evenness index of gut microbiota at 75 days.
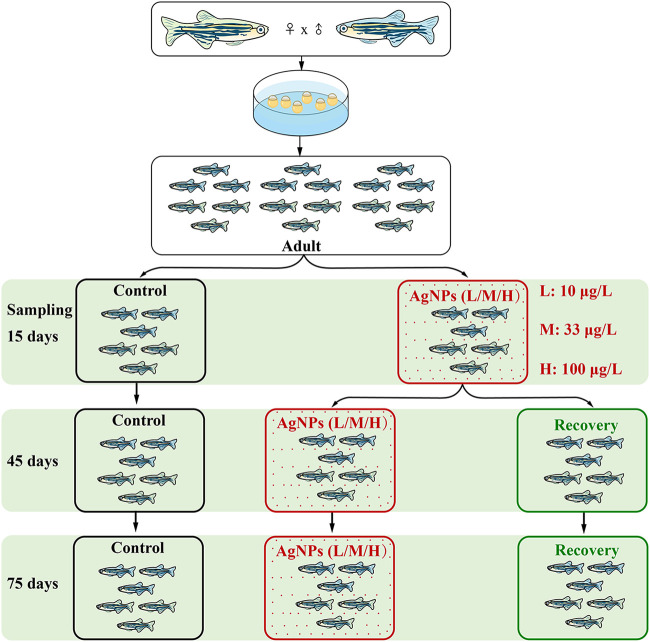
FIG 1.
Experimental design. The resilience and resistance of gut microbiota were investigated using the same batch of adult zebrafish, which were subjected to exposure to three concentrations of AgNPs (i.e, 10, 33, and 100 μg/liter) for 15 days. They were then randomly assigned to continued-exposure (red) or recovery (green) groups for 30 and 60 days, respectively. The continued-exposure and recovery groups were used to test microbial resistance and resilience, respectively. The controls (black) without AgNP exposure were also sampled at each sampling time. L, M, and H indicate exposure to low (10 μg/liter), medium (33 μg/liter), and high (100 μg/liter) concentrations of AgNPs, respectively.
FIG 2.
Box plot of Shannon index (A, D, G), observed OTUs (B, E, H), and Pielou evenness index (C, F, I) of gut microbiota in zebrafish after AgNP exposure. Different lowercase letters indicate significant differences (ANOVA, P < 0.05) among groups. C, control; L, M, and H, exposure to low (10 μg/liter), medium (33 μg/liter), and high (100 μg/liter) concentrations of AgNPs, respectively; L-R, M-R, and H-R, recovery from exposure to low (10 μg/liter), medium (33 μg/liter), and high (100 μg/liter) concentrations of AgNPs, respectively. Data are the mean ± standard error (n = 6).
Principal-coordinate analysis (PCoA) demonstrated that gut microbial structure after 15 days of AgNP disturbance was considerably different from that of controls, but it gradually returned to its normal state after 75 days of recovery (Fig. 3). Although there have some variations among replicates, they do not change the observed trend. With the AgNP concentration increasing, three exposure groups were clearly separated from the controls after 15 days of exposure (Fig. 3A). At 45 days, exposure and recovery groups were almost separated from the controls by the PCoA (Fig. 3B). Surprisingly, given our expectation that AgNP exposure would affect community recovery, a certain similarity of community structure between the recovery groups and controls was detected among gut microbial communities at 75 days (Fig. 3C). So, longer recovery time decreased the difference in microbial structure among groups. These community patterns could be further confirmed by the nonparametric tests (i.e., multiple-response permutation procedure [MRPP], permutational multivariate analysis of variance [PERMANOVA], and analysis of similarity [ANOSIM]) based on the Bray-Curtis distance, which showed significant (P < 0.05) differences among the gut microbial communities for each pairwise comparison (Table 1).
FIG 3.
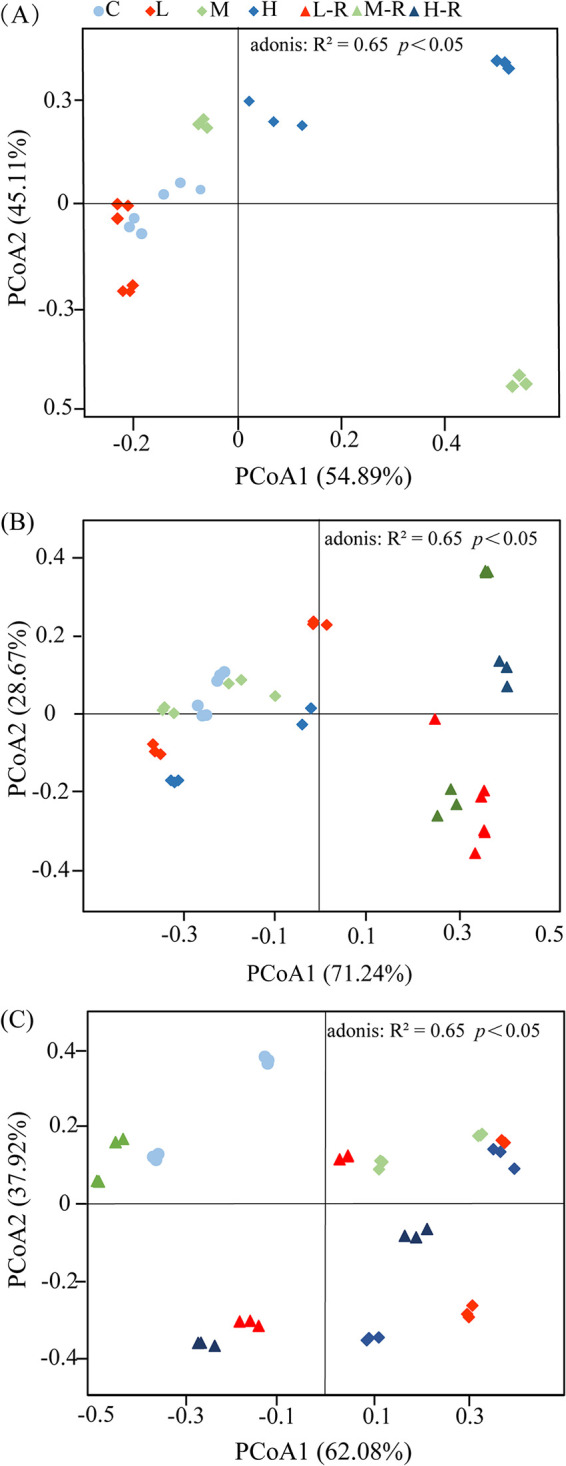
Principal-coordinate analysis (PCoA) of the gut microbiota of zebrafish subjected to AgNP treatments of 15 days (A), 45 days (B), and 75 days (C). C, control; L, M, and H, exposure to low (10 μg/liter), medium (33 μg/liter), and high (100 μg/liter) concentrations of AgNPs, respectively; L-R, M-R, and H-R, recovery from exposure to low (10 μg/liter), medium (33 μg/liter), and high (100 μg/liter) concentrations of AgNPs, respectively.
TABLE 1.
Dissimilarity tests of gut microbial communities based on three nonparametric testsa
| Sampling time and group comparison | MRPP |
ANOSIM |
PERMANOVA |
|||
|---|---|---|---|---|---|---|
| Delta | P | R | P | F | P | |
| 15 days | ||||||
| C vs L | 0.253 | 0.001 | 0.850 | 0.004 | 14.153 | 0.004 |
| C vs M | 0.364 | 0.003 | 0.495 | 0.002 | 6.680 | 0.002 |
| C vs H | 0.340 | 0.008 | 0.588 | 0.003 | 11.365 | 0.004 |
| 45 days | ||||||
| C vs L | 0.328 | 0.002 | 0.909 | 0.003 | 18.338 | 0.004 |
| C vs M | 0.283 | 0.004 | 0.646 | 0.001 | 4.412 | 0.003 |
| C vs H | 0.267 | 0.005 | 0.938 | 0.004 | 11.380 | 0.002 |
| C vs L-R | 0.305 | 0.003 | 0.892 | 0.002 | 9.5674 | 0.007 |
| C vs M-R | 0.362 | 0.004 | 0.888 | 0.002 | 13.091 | 0.003 |
| C vs H-R | 0.234 | 0.020 | 1.000 | 0.0.013 | 33.933 | 0.010 |
| 75 days | ||||||
| C vs L | 0.352 | 0.002 | 1.000 | 0.003 | 17.957 | 0.003 |
| C vs M | 0.310 | 0.006 | 0.892 | 0.002 | 16.502 | 0.002 |
| C vs H | 0.423 | 0.001 | 1.000 | 0.004 | 11.714 | 0.003 |
| C vs L-R | 0.404 | 0.005 | 0.585 | 0.003 | 7.777 | 0.003 |
| C vs M-R | 0.361 | 0.002 | 0.827 | 0.003 | 11.587 | 0.001 |
| C vs H-R | 0.437 | 0.004 | 0.580 | 0.003 | 7.167 | 0.004 |
C, control; L, M, and H, exposure to treatments of low (10 μg/liter), medium (33 μg/liter), and high (100 μg/liter) concentrations of AgNPs, respectively; L-R, M-R, and H-R, recovery from exposure to low (10 μg/liter), medium (33 μg/liter), and high (100 μg/liter) concentrations of AgNPs, respectively; MRPP, multiple-response permutation procedure; ANOSIM, analysis of similarities; PERMANOVA, permutational multivariate analysis of variance; Delta, a measure of dissimilarity by means of a “delta score”; R, the correlation coefficient, where an R value near +1 means that there is dissimilarity between the groups, while an R value near 0 indicates no significant dissimilarity between the groups; F, pseudo-f statistic for testing the null hypothesis.
Community composition in relation to resistance and resilience.
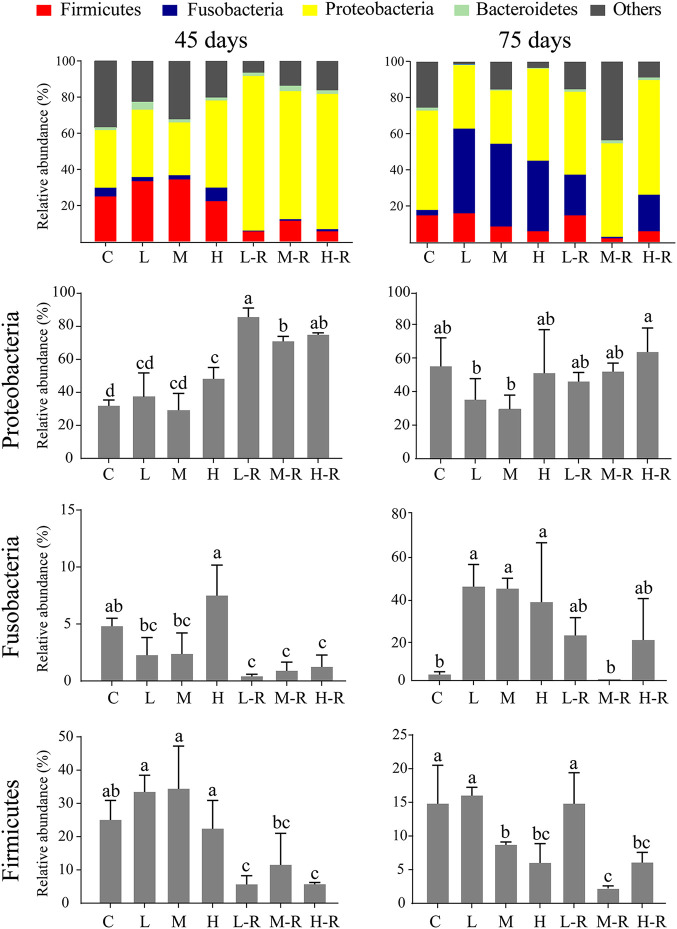
Zebrafish gut microbial communities were generally dominated by the phyla Firmicutes, Fusobacteria, Proteobacteria, and Bacteroidetes (Fig. 4; see Fig. S1 in the supplemental material). At 45 days, no significant changes were observed between exposure groups and the controls at the phylum level, but Proteobacteria detected in the high-concentration exposure group were significantly (P < 0.05) increased. In contrast, the relative abundance of Proteobacteria in all the recovery groups was significantly higher than that in the control and exposure groups (P < 0.05). In addition, the relative abundance of Fusobacteria in the recovery groups ranged from 0.4% to 1.2%, significantly lower than that of the controls. The relative abundance of Firmicutes in the recovery groups ranged from 5.6% to 11.5%. However, the relative abundance of Proteobacteria at 45 days was negatively correlated with that of Fusobacteria and Firmicutes, and the relative abundance of Proteobacteria and Fusobacteria in the recovery groups was not significantly different from that of controls at 75 days.
FIG 4.
Relative abundances of the dominant phyla of zebrafish gut microbiota at 45 days (left panel) and 75 days (right panel). Different lowercase letters above the bars indicate significant differences among groups (ANOVA, P < 0.05), whereas the same letter suggests no significant difference. Data are the mean ± standard error (n = 6).
Relative abundance of dominant phyla of zebrafish gut microbiota at 15 days of exposure. Different letters above the bars indicate significant differences among groups (ANOVA, P < 0.05), whereas the same letter suggests no significant difference. Download FIG S1, TIF file, 0.2 MB (156.4KB, tif) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
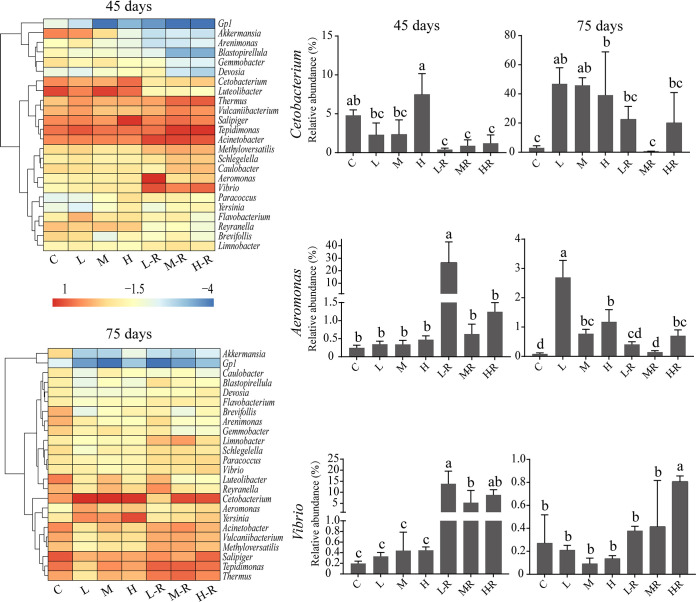
The heatmap with hierarchical clustering of the top 25 genera (Fig. 5) indicated that the most abundant genera across all samples were Cetobacterium, Tepidimonas, Acinetobacter, Thermus, Vulcaniibacterium, Salipiger, Luteolibacter, Methyloversatilis, Paracoccus, Vibrio, and Aeromonas. At 45 days, the percentage of Cetobacterium ranged from 0.9% to 7.5%, while it ranged from 0.7% to 47.0% at 75 days and showed no significant differences from that of the controls in the recovery groups. The abundance of Aeromonas detected in the recovery groups showed significant differences from that in the controls at 45 days, and the L-R and M-R groups showed no significant differences from the controls at 75 days. Similarly, the relative abundance of Vibrio in the L-R and M-R groups was not significantly different from that of the controls at 75 days. Among the recovery groups, some genera increased their relative abundances, such as Acinetobacter (from 4.8% to 15.6%), Tepidimonas (from 7.2% to 15.4%), and Thermus (from 1.2% to 6.7%). In contrast, the abundances of other genera decreased after exposure. For example, Akkermansia decreased from 5.2% to 1.6% and Gemmobacter decreased from 0.67% to 0.27% in the exposure groups. The relative abundance of Cetobacterium in the 75-day exposure groups was positively correlated with that of Aeromonas, while the relative abundance of Aeromonas in the recovery groups was negatively correlated with that of Vibrio. Overall, gut microbial composition dynamics within each treatment during the recovery period revealed only some minor differences.
FIG 5.
Relative abundances of the dominant genera of zebrafish gut microbiota. Different lowercase letters above the bars indicate significant differences among groups (ANOVA, P < 0.05), whereas the same letter suggests no significant difference. Data are the mean ± standard error (n = 6).
To better understand the more refined variations among different treatments, we also compared the variations of “core” OTUs (35), which are defined as those detected in over 80% of samples with an abundance of >1% for each sample of the considered groups (Table S1). OTU_1 (Cetobacterium), OTU_2 (Firmicutes), OTU_3 (Tepidimonas), OTU_4 (Rhodobacteraceae), and OTU_5 (Thermus) were identified as core taxa in all samples. Interestingly, the opportunistic pathogen of OTU_7 (Citrobacter) was significantly enriched in the recovery groups at 75 days. At 45 days, 11, 10, and 12 of the core OTUs detected in control, exposure, and recovery groups accounted for 75.8%, 73.8%, and 77.6% of the total abundance in each group. At 75 days, 17, 11, and 14 core OTUs from the control, exposure, and recovery groups accounted for 73.9%, 88.5%, and 69.1% of the total abundances of each group (Fig. S2). Thus, both gut microbial community composition and the abundance of core OTUs in zebrafish gut showed clear dynamics.
Core OTUs (shared by more than 80% of the samples and with a relative abundance of >1%) for different treatments. The relative abundances (A) and a Venn diagram showing the number of shared and unique core OTUs (B) in different treatments are shown. Download FIG S2, TIF file, 1.0 MB (1MB, tif) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Taxonomy of each core OTU (shared by >80% samples and with abundances of >1%) at two sampling times (45 days and 75 days). Download Table S1, DOCX file, 0.02 MB (20.1KB, docx) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Ecological processes for assessing resistance and resilience.
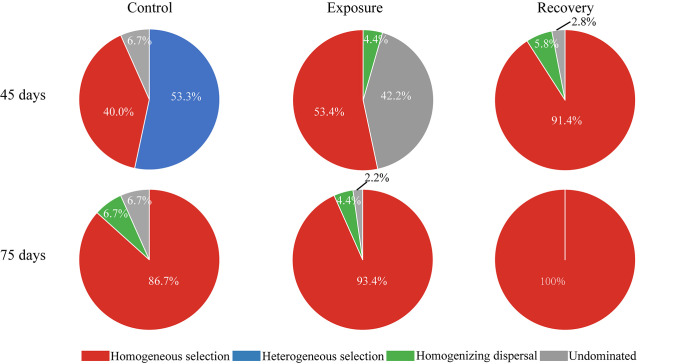
To clarify the ecological mechanisms underlying the resistance and resilience of microbial community assembly, we further quantified the major ecological processes. We found that the resistance and resilience of gut microbiota were governed by strong deterministic selection processes (Fig. 6). At 45 days, the proportions of deterministic selection (53.4%) and undominated (42.2%) processes were similar in the exposure groups, while the deterministic process was the leading force for microbial assembly in controls (93.4%) and recovery groups (91.4%). At 75 days, deterministic selection was also the major force for microbial assembly in controls (86.7%), exposure groups (93.4%), and recovery groups (100%). Specifically, homogeneous selection, which causes community composition to be similar under consistent environmental conditions, was responsible for 40.0% to 100% of the gut microbial variations and increased with the recovery time. In contrast, there was only weak (4.4%) homogenizing dispersal in the exposure groups, but the contribution of the undominated process was also important (42.2%) throughout the resistance of gut microbiota. Thus, the resilience and resistance of zebrafish gut microbiota appeared to be governed by a deterministic process, especially homogeneous selection in the exposure and recovery groups.
FIG 6.
Contribution of ecological processes to the assembly of zebrafish gut microbiota among different treatments.
Networks and interactions to evaluate resistance and resilience.
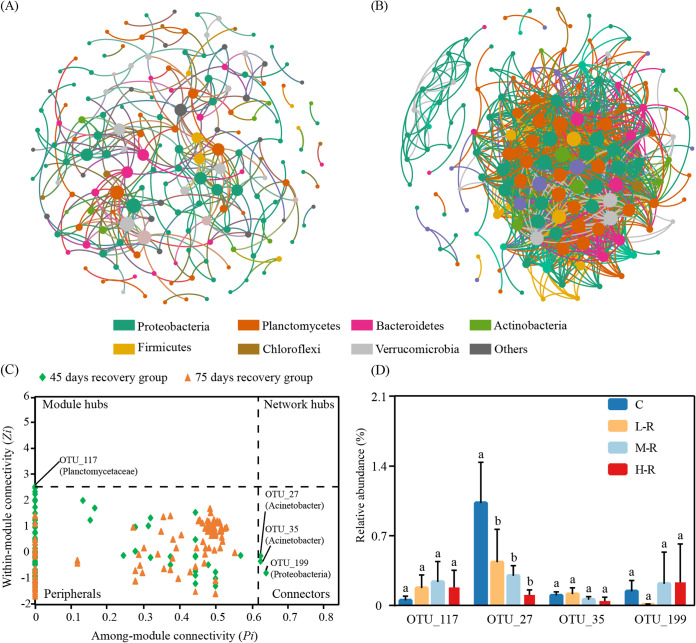
In order to evaluate resistance and resilience with respect to the microbial interactions within zebrafish gut microbiota, correlation-based networks were constructed (Fig. 7A and B; Fig. S3A and B). At 45 days, the community interactions of gut microbiota showed a simple network (average connectivity [avgK], 3.475) but with the highest positive connections (82.0%) in the recovery groups (Table 2). The observed changes were increased with increasing recovery time. The network complexity (a higher average degree representing a greater network complexity) at 75 days for the recovery groups was higher than that at 45 days, but the average path distance (2.161) was slightly reduced. Multiple network topological metrics consistently suggested that networks of treatment groups at 75 days were more complex than those at 45 days. Additionally, we visualized the modules with at least two nodes for further analysis (Fig. S4). The positive links ranged from 62.7% to 93.8%, and the network modularity was >0.174 in all networks (Table 2).
FIG 7.
Correlation-based networks of zebrafish gut microbial OTUs in the 45-day (A) and 75-day (B) recovery groups. Zi-Pi plots show the distribution of OTUs based on their topological roles (C) and the abundance of potential keystone OTUs (D). Each node represents an OTU, and threshold values of Zi and Pi for categorizing OTUs were 2.5 and 0.62, respectively.
TABLE 2.
Topological properties of empirical molecular ecological networks in comparison to random networks
| Sampling time and group | Total no. of nodes | Empirical networksa |
Random networksa,b |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total no. of links | No. of positive links | avgK | avgCC | GD | Modularity | Nodes with maximum stress centrality | Nodes with maximum eigenvector centrality | Nodes with maximum betweenness | avgCC | GD | ||
| 45 days | ||||||||||||
| Control | 232 | 564 | 408 | 4.862 | 0.382 | 8.274 | 0.766 | OTU_115 | OTU_12 | OTU_115 | 0.056 ± 0.009 | 3.419 ± 0.044 |
| Exposure | 257 | 643 | 486 | 5.004 | 0.283 | 4.311 | 0.690 | OTU_2 | OTU_5 | OTU_2 | 0.691 ± 0.008 | 1.526 ± 0.002 |
| Recovery | 179 | 311 | 255 | 3.475 | 0.285 | 6.335 | 0.740 | OTU_59 | OTU_3 | OTU_59 | 0.033 ± 0.010 | 3.856 ± 0.083 |
| 75 days | ||||||||||||
| Control | 167 | 1,236 | 766 | 12.026 | 0.526 | 3.442 | 0.479 | OTU_109 | OTU_87 | OTU_109 | 0.155 ± 0.010 | 2.748 ± 0.026 |
| Exposure | 304 | 1,828 | 1,681 | 14.802 | 0.471 | 4.896 | 0.174 | OTU_145 | OTU_131 OTU_323 |
OTU_123 | 0.227 ± 0.010 | 2.413 ± 0.026 |
| Recovery | 146 | 1,704 | 1,584 | 23.342 | 0.613 | 2.161 | 0.411 | OTU_96 | OTU_24 | OTU_96 | 0.523 ± 0.015 | 2.166 ± 0.025 |
avgK, average connectivity; avgCC, average clustering coefficient; GD, average path distance.
Values are the mean ± standard deviation generated from networks randomly rewired 100 times.
(A and B) Correlation-based network of zebrafish gut microbial OTUs detected in groups at 45 days (A) and 75 days (B) of exposure. Each node corresponds to an OTU. (C) Zi-Pi plots show the distribution of OTUs based on their topological roles in two exposure groups. (D) Abundance of keystone OTUs. Threshold values of Zi and Pi for categorizing OTUs were 2.5 and 0.62, respectively. Download FIG S3, TIF file, 2.3 MB (2.3MB, tif) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Highly connected modules within zebrafish gut microbial networks for different treatments (control, exposure, and recovery). Colors of nodes indicate different major phyla. Red links indicate positive correlations between two individual nodes, whereas blue links indicate negative correlations between nodes. Download FIG S4, TIF file, 2.9 MB (2.9MB, tif) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The possible topological roles of nodes in the gut microbiota were determined by their within-module connectivity (Zi) and among-module connectivity (Pi). There were three connectors (OTU_27, OTU_35, and OTU_199) for the recovery groups at 45 days, but there was no connector for the recovery groups at 75 days (Fig. 7C). Similarly, only one module hub (OTU_117) was detected for the recovery groups at 45 days. The OTU_27 (Acinetobacter) with 0.5% relative abundance showed significant differences from that of the controls at 45 days (Fig. 7D). We also evaluated the contributions of the potential keystone taxa to the exposure groups for resistance. The potential keystone taxa in exposure groups at 45 and 75 days were mainly from Planctomycetes (including OTU_54, OTU_73, OTU_124, OTU_235, and OTU_390). Moreover, the relative abundances of OTU_124 (0.6%) (Planctopirus), OTU_73 (0.9%) (Gemmata), and OTU_54 (0.9%) (Planctomycetaceae) were significant different from that of the controls (Fig. S3C and D). Our results showed that AgNP exposure affected the network structure and topological roles of individual OTUs and the potential keystone taxa.
DISCUSSION
Understanding ecological stability by characterizing community resilience and resistance to disturbances is crucial for predicting microbial responses to environmental changes. However, such knowledge is very limited for animal gut microbiota, which was recently considered to be an extra “organ” of the host (36). We found that the gut microbiota in zebrafish subjected to AgNP exposure was pushed toward an alternative state after recovery of the water environment. Notably, maintenance of the stability of the zebrafish gut microbiota might be attributed mainly to the cooperation of some beneficial microbes, and homogeneous selection was one of the major ecological processes driving gut microbiota recovery after AgNP disturbance. These results supported our hypothesis that gut microbial resilience and resistance were governed by selecting an adaptive taxonomic composition and increasing their microbial interactions in response to environmental disturbances.
Resilience and resistance are inherent properties to characterize the stability of microbial communities, which cannot be independent of the community diversity and composition (37). Generally, higher microbial diversity always shows stronger resistance to continuous disturbances and might be easier to recover after the disturbance is excluded (38). We found that continued AgNP exposure of zebrafish decreased both the diversity and richness of gut microbiota at 15 and 75 days. This finding is similar to that reported in exposure of fathead minnows to a low level of triclosan (39). Moreover, the increasing diversity in medium concentration of exposure and recovery groups indicated that moderate disturbances would destabilize the microbial niches (40). These niches were colonized by new species and thus resulted in increasing diversity (41). We know that the ecotoxicity of nanoparticles could suppress sensitive gut microbes (42) and promote pollutant-tolerant members (e.g., Proteobacteria), which may significantly affect the composition of gut microbiota in zebrafish exposed to AgNPs. This was confirmed by the community patterns at 15 and 75 days, as visualized in PCoA ordination. The disturbed microbial community could, after a certain time of environmental recovery, rebound to a state comparable to that before disturbance but not to a complete recovery. This result was identical to that of human microbiomes perturbed with antibiotic exposure (43). On the other hand, a convergent trend of microbial patterns between the exposure groups and controls at 45 days indicated that the gut microbiota also could resist some of the AgNP disturbance.
The variations of specific microbial taxa in the zebrafish gut may also affect gut stability after AgNP disturbance. For example, the rapid growth of Proteobacteria, which is considered a potential diagnostic signature of dysbiosis and disease risk, may induce the gut microbiota to an unstable state (44, 45). The higher abundance of Proteobacteria was also a typical feature of zebrafish with inflammatory bowel disease induced by environmental pollutants (46). In contrast, the cooperation of beneficial microbes such as Fusobacteria, Bacteroidetes, and Firmicutes could result in gut stability by producing essential vitamins to assist in detoxification (47). The significant differences of particular genera between the recovery groups and controls also suggested an incomplete recovery. For example, there was a significant increase of fish pathogens (e.g., Vibrio and Aeromonas) in the recovery groups. The appearance of specific core OTUs in the gut may be partly due to a different selective pressure within the host gut habitat (48). AgNP disturbance can facilitate the proliferation of opportunistic pathogens (e.g., core OTU_7 [Citrobacter]), consistent with other studies of fish microbiota recovery (49). Thus, changes in abundance of microbial taxa at different levels all represented direct resistant and resilient responses to AgNP exposure.
Our findings also revealed the roles of ecological processes affecting the resistance and resilience of gut microbiota to AgNP exposure. Dispersal and colonization are two stochastic forces that lead to unpredictable variability in community composition (50–52). This study indicated that homogenizing dispersal was important only in exposure groups, with the explanation that the microbial diversity showed no significant decrease after 45 days of AgNP exposure. We found that the deterministic process of homogeneous selection governed over 53% of community turnover. That means that the responses of gut microbiota to AgNP disturbance could be explained by the insurance hypothesis, as the taxonomic composition is resilient to disturbances by increasing abundance and compensating for functions previously carried by sensitive taxa. This finding is consistent with a previous study that suggested that the relative importance of selection contributed to microbial resilience by accelerating the adaptation of communities (9). This may be partly due to the rapid growth of Cetobacterium, as it can quickly adapt to suitable host conditions (53). Of course, some other processes, such as microbial interactions and active dispersal currently classified as the undominated process, may also facilitate the resilience and resistance of fish gut microbiota.
However, neither composition nor ecological processes can demonstrate the microbial interactions to address the resistance and resilience of gut microbiota. Network analyses were applied to illustrate potential microbial community interactions and explore the resistance of native taxa (54, 55). The higher avgCC detected herein revealed a highly complex and modular microbiome, which might resist environmental disturbances more efficiently. A recent study found that complex microbial networks could increase intestinal microbial stability in Atlantic salmon after exposure to antibiotics (56). Therefore, the microbial resilience and resistance might increase the stability of the cooccurrence network of microbial communities in zebrafish subjected to AgNP exposure. Nevertheless, the stability of the network did not recover to its original level in the recovery group. Moreover, increasing positive interactions were observed in the AgNP exposure groups, indicating an enhancement of community stability. Evidence has suggested that a moderate disturbance would enhance the complexity of microbial interactions (57). In mutualistic ecosystems, beneficial microbes have evolved a distinct defense strategy to protect against pathogens and maintain community stability (58, 59). One previous study suggested that increasing cooperative relationships within microbe interactions may help them be more tolerant of disturbances (41). Collectively, cooperative relationships and complex microbial interactions of zebrafish gut microbiota represented good resistance and resilience to AgNP disturbance.
Also, highly connected taxa have been proposed to be potential keystone taxa, which may play a vital role in restoring a dysbiosis community to a predisturbance composition (60). The concentrated keystone taxa suggest that they may maintain the stability of the gut microbiota at 45 days. We found that the keystone taxa occupied a great proportional influence in the intestinal microbiota, and their absence may lead to network fragmentation in the treatment groups (61). For example, the representative keystone genus in the recovery process is Acinetobacter, which might positively affect the digestive processes of fish because of enzyme production (62). Similarly, the keystone genus of Gemmata has been identified to have a versatile hydrolytic capability and a high ability to decompose organic matters by relieving the digestive pressure of intestines to enhance the process of resistance (63). Furthermore, due to the lack of peptidoglycan in the cell wall of Planctomycetes, they are able to resist harsh environments and become the keystone of the process of resistance (64). As a result, our attempts exemplified that the stable keystone taxa in the cooccurrence patterns of gut bacterial assembly contributed to gut microbial resilience and resistance.
In conclusion, the AgNP exposure of zebrafish resulted in gut microbiota changes and reduced its α-diversity. We also found that AgNP exposure pushed gut microbial communities toward an alternative state that could facilitate the proliferation of opportunistic pathogens (e.g., Citrobacter). The resilience of zebrafish gut microbiota responses to AgNP disturbances might be correlated with selecting an adaptive taxonomic composition and increasing key taxonomic interactions. Additionally, our results indicated that the microbial community resistance and recovery were governed mainly by a deterministic selection process. This ecological resilience and resistance of the host-associated microbiome increased our understanding of environmental disturbance implications for fish gut ecology.
MATERIALS AND METHODS
AgNP characterization and solution preparation.
AgNPs (99%; CAS no. 576832-5G) used in this experiment were purchased from Sigma-Aldrich (St. Louis, MO, USA). Although the diameter of the AgNPs was suggested to be approximately 100 nm, we measured their size distribution by dynamic light scattering (DLS) using a Malvern Zetasizer Nano ZS in ultrapure water (100 μg/liter). The size and morphology of AgNPs were characterized by transmission electron microscopy (TEM) and X-ray diffraction (XRD) (see Fig. S5 in the supplemental material). For the stock preparation, AgNP powder was suspended in ultrapure water (Millipore, Billerica, MA) at a concentration of 100 mg/liter. To maintain a stable AgNP concentration, the initial suspension was sonicated (50 W/liter, 40 kHz) for 50 min at 60°C. The stock solution was diluted before use with fresh charcoal-filtered water and resonicated (50 W/liter, 40 kHz). The AgNP stock solutions were then prepared every 48 h to maintain relatively consistent exposure levels. Finally, half of the exposure water was refreshed daily, and the entire exposure water was refreshed weekly to keep consistent concentrations of AgNPs in each tank.
Characteristics of AgNPs. (A) AgNP image taken by a transmission electron microscope (TEM). (B) Particle size dispersion of the AgNPs measured by dynamic light scattering (DLS). (C) X-ray diffraction (XRD) patterns of AgNPs. FIG S5, TIF file, 1.4 MB (1.4MB, tif)
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Experimental setup and zebrafish husbandry.
To investigate the ecological resistance and resilience of the gut microbial community, we monitored the succession of gut microbiota in experimental zebrafish (wild type) subjected to the disturbance of AgNP exposure. Adult zebrafish (AB strain) were reared using glass tanks and fed newly hatched brine shrimp (Artemia nauplii) twice daily. Specifically, fully aerated tap water was used during the experiment, and relatively stable conditions (28°C ± 0.5°C and a 14-h light/10-h dark cycle) were applied to avoid additional environmental perturbations. The adult zebrafish were exposed to three different concentrations of AgNPs (i.e., 10, 33, and 100 μg/liter), accompanied by a control group without AgNP exposure. After the zebrafish were exposed to different concentrations of AgNPs for 15 days, they were randomly assigned to continued exposure (Fig. 1, indicated by red) and recovery (Fig. 1, indicated by green) groups. The experiment was then extended an additional 2 months after 15 days of exposure to test the resistance and resilience of the gut microbiota.
Sampling procedures and DNA extraction.
At each sampling time point (i.e., 15, 45, and 75 days), three female and three male zebrafish were randomly collected from each group (Fig. 1) for gut microbial analysis by sequencing the 16S rRNA gene. However, sex factor was not considered in further analysis due to no statistical differences among females and males (data no shown). The whole intestine of each zebrafish was immediately removed aseptically as previously described (31), and each intestine was regarded as a single sample and stored in a sterile 1.5-ml tube at −80°C until DNA extraction. In total, 105 zebrafish gut samples were obtained for the following DNA extraction. All protocols involved in the animal experiments were approved by the Institutional Animal Care and Use Committee of the Institute of Hydrobiology, Chinese Academy of Sciences (approval identifier Keshuizhuan 08529).
Genomic DNA was extracted using the PowerFecal (gut samples) DNA isolation kit (Mo Bio, CA, USA) in accordance with the manufacturer’s instructions. The concentrations and quality of extracted DNA were determined using a NanoDrop One spectrophotometer (Thermo Fisher Scientific, MA, USA), and all samples were diluted to the same concentration (10 ng/μl) for subsequent PCR amplification.
Sequencing analysis of the 16S rRNA gene.
The V4 region of the 16S rRNA gene was amplified by the primer set 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). Each sample was amplified in a reaction volume of 50 μl containing 1× Premix Taq DNA polymerase (buffer, deoxynucleoside triphosphate [dNTP], and Taq were included), 0.2 mM each primer, and 50 ng genomic DNA by using the following procedures: predenaturation for 5 min at 95°C and then 30 cycles of 30 s at 95°C, 30 s at 52°C, and 30 s at 72°C, followed by a postextension of 10 min at 72°C. Negative-control experiments were always performed to ensure that no contamination had occurred. PCR products were visualized using 1% agarose gels stained with ethidium bromide. After all samples were successfully amplified, the PCR products of each sample were quantified and equally combined. The target band visualized by 2.0% agarose gel was excised and purified with a QIAquick gel extraction kit (Qiagen, CA, USA). After requantification of the concentration of the purified DNA, it was subjected to library construction. The constructed amplicon library was finally sequenced by the Illumina HiSeq 2500 platform (Illumina, CA, USA) at Guangdong Magigene Biotechnology Co., Ltd., with a 2 × 250 bp kit.
Quality filtering and processing of sequence reads were conducted using the publicly available Galaxy pipeline (http://mem.rcees.ac.cn:8080/) as described previously (65). In brief, overlapped paired-end sequences were first assembled using FLASH (66), and poorly overlapped and low-quality sequences such as those with a length of <140 bp and a moving-window (5 bp) quality score of <20 were removed before downstream analysis. After removal of chimeras, UPARSE was used to classify high-quality sequences into operational taxonomy units (OTUs) at a cutoff of 97% identity. To make the sequencing depth the same for all samples, all 105 samples were rarefied to 14,825 reads per sample for subsequent analyses.
Ecological process analysis.
To quantify the ecological processes governing the gut microbial community, we calculated the major ecological processes as previously described (30, 65). In this framework, the variation or turnover of both phylogenetic diversity and taxonomic diversity was first measured with the null model-based phylogenetic and taxonomic β-diversity metrics. The β-nearest taxon indices (β-NTI) and Raup-Crick index (RCBray) were then used to quantify the ecological processes that influence gut microbial community composition on a spatiotemporal scale. The community turnover is governed by heterogeneous (β-NTI > 2) or homogeneous (β-NTI less than −2) selection. Pairwise comparisons with a |β-NTI| of <2 were further subjected to RCBray: the fraction of pairwise comparisons with a |β-NTI| of <2 and an RCBray of less than −0.95 estimated the homogenizing dispersal influence; the fraction of pairwise comparisons with a |β-NTI| of <2 and an RCBray of >0.95 estimated the dispersal limitation influence; and the remaining fraction of pairwise comparisons with a |β-NTI| of <2 and an |RCBray| of <0.95 represented the component of compositional turnover undominated by any process mentioned above (67).
Molecular ecological network construction.
To reveal the gut microbial interactions in response to AgNP disturbance, we constructed networks with the publicly available Molecular Ecological Network Analysis Pipeline (MENA; http://ieg2.ou.edu/MENA/) (17) based on the OTU relative abundances. Covariations were measured across 6 to 18 biological replicates for each network, and only OTUs detected in more than two-thirds of the samples of each group were kept in network construction. Random matrix theory (RMT) was used to automatically identify the appropriate similarity threshold (St) prior to network construction (68), but all the networks were constructed using the same St (i.e., 0.92). Two topological parameters estimated the roles of individual nodes (OTUs) in the network: the within-module connectivity, Zi, which quantified to what extent a node connected to other nodes in its module, and the among-module connectivity, Pi, which quantified how well the node connected to different modules. The nodes with a high value of either Zi or Pi were defined as potential keystone taxa, including module hubs (Zi > 2.5, Pi ≤ 0.62; critical to the module coherence), connectors (Zi ≤ 2.5, Pi > 0.62; connect modules together and important to network coherence), and network hubs (Zi > 2.5, Pi > 0.62; vital to both the network and its own module coherence). The networks were graphed using Gephi 0.9.2 and Cytoscape 3.8.2.
Statistical analysis.
The comparisons of α- and β-diversities were used to indicate changes in the gut microbiota in response to AgNP disturbances. The indices of Shannon, Pielou’s evenness, and observed OTUs were calculated using the VEGAN package (v.2.3.5) in R (v.3.4.4). In order to elucidate whole patterns of microbial communities, Bray-Curtis distance was used to reveal community structure based on principal-coordinate analysis (PCoA). A heatmap was constructed between the relative abundances of dominant OTUs using the AUTOMAP package. The multiple-response permutation procedure (MRPP), permutational multivariate analysis of variance (PERMANOVA), and analysis of similarity (ANOSIM) tests were performed to test the community dissimilarity by the VEGAN package in R (version 3.4.4) (69). Significance tests were performed using one-way analysis of variance (ANOVA) with Tukey’s test by using GraphPad Prism 8 (GraphPad Software, Inc., San Diego, CA, USA). A P value of <0.05 indicated significant differences, and all values are presented as the mean ± standard error of the replicates in each group.
Data availability.
The raw sequencing data are available at the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) under accession number PRJNA668536.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant no. 31802350 and 92051120), the Innovation Group Project of Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai) (grant no. 311021006), the Fundamental Research Funds for the Central Universities (grant no. 19lgzd28), the Hundred Talents Program through Sun Yat-sen University (grant no. 18821107), and the Youth Innovation Promotion Association of the Chinese Academy of Sciences (grant no. 2019333).
Y.Y., Q.Y., and J.H. designed the study. L.R. and W.Z. conducted the experiments. P.C. and F.X. analyzed the data. P.C. wrote the first draft of the manuscript. F.X., X.C., H.Y., Y.W., K.X., X.Z., R.H., Z.H., and Q.Y. contributed substantially to manuscript writing, editing, or revisions.
Contributor Information
Qingyun Yan, Email: yanqy6@mail.sysu.edu.cn.
Rosie Alegado, University of Hawaii at Manoa.
REFERENCES
- 1.Shade A, Read JS, Youngblut ND, Fierer N, Knight R, Kratz TK, Lottig NR, Roden EE, Stanley EH, Stombaugh J, Whitaker RJ, Wu C, McMahon KD. 2012. Lake microbial communities are resilient after a whole-ecosystem disturbance. ISME J 6:2153–2167. doi: 10.1038/ismej.2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griffiths BS, Philippot L. 2013. Insights into the resistance and resilience of the soil microbial community. FEMS Microbiol Rev 37:112–129. doi: 10.1111/j.1574-6976.2012.00343.x. [DOI] [PubMed] [Google Scholar]
- 3.Philippot L, Griffiths BS, Langenheder S. 2021. Microbial community resilience across ecosystems and multiple disturbances. Microbiol Mol Biol Rev 85:e00026-20. doi: 10.1128/MMBR.00026-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo X, Gao Q, Yuan M, Wang G, Zhou X, Feng J, Shi Z, Hale L, Wu L, Zhou A, Tian R, Liu F, Wu B, Chen L, Jung CG, Niu S, Li D, Xu X, Jiang L, Escalas A, Wu L, He Z, van Nostrand JD, Ning D, Liu X, Yang Y, Schuur EAG, Konstantinidis KT, Cole JR, Penton CR, Luo Y, Tiedje JM, Zhou J. 2020. Gene-informed decomposition model predicts lower soil carbon loss due to persistent microbial adaptation to warming. Nat Commun 11:4897. doi: 10.1038/s41467-020-18706-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Svensson JR, Lindegarth M, Pavia H. 2009. Equal rates of disturbance cause different patterns of diversity. Ecology 90:496–505. doi: 10.1890/07-1628.1. [DOI] [PubMed] [Google Scholar]
- 6.Allison SD, Martiny JB. 2008. Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci USA 105(Suppl 1):11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou J, Deng Y, Zhang P, Xue K, Liang Y, Van Nostrand JD, Yang Y, He Z, Wu L, Stahl DA, Hazen TC, Tiedje JM, Arkin AP. 2014. Stochasticity, succession, and environmental perturbations in a fluidic ecosystem. Proc Natl Acad Sci USA 111:E836–E845. doi: 10.1073/pnas.1324044111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cadotte MW, Davies TJ, Regetz J, Kembel SW, Cleland E, Oakley TH. 2010. Phylogenetic diversity metrics for ecological communities: integrating species richness, abundance and evolutionary history. Ecol Lett 13:96–105. doi: 10.1111/j.1461-0248.2009.01405.x. [DOI] [PubMed] [Google Scholar]
- 9.Heuer H, Smalla K. 2012. Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol Rev 36:1083–1104. doi: 10.1111/j.1574-6976.2012.00337.x. [DOI] [PubMed] [Google Scholar]
- 10.Tesson SV, Okamura B, Dudaniec RY, Vyverman W, Löndahl J, Rushing C, Valentini A, Green AJ. 2015. Integrating microorganism and macroorganism dispersal: modes, techniques and challenges with particular focus on co-dispersal. Ecoscience 22:109–124. doi: 10.1080/11956860.2016.1148458. [DOI] [Google Scholar]
- 11.Székely AJ, Langenheder S. 2017. Dispersal timing and drought history influence the response of bacterioplankton to drying-rewetting stress. ISME J 11:1764–1776. doi: 10.1038/ismej.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JB. 2012. Beyond biogeographic patterns: processes shaping the microbial landscape. Nat Rev Microbiol 10:497–506. doi: 10.1038/nrmicro2795. [DOI] [PubMed] [Google Scholar]
- 13.Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C. 2012. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol 8:e1002606. doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, FitzGerald MG, Fulton RS. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Relman DA. 2012. The human microbiome: ecosystem resilience and health. Nutr Rev 70(Suppl 1):S2–S9. doi: 10.1111/j.1753-4887.2012.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steele JA, Countway PD, Xia L, Vigil PD, Beman JM, Kim DY, Chow C-ET, Sachdeva R, Jones AC, Schwalbach MS, Rose JM, Hewson I, Patel A, Sun F, Caron DA, Fuhrman JA. 2011. Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J 5:1414–1425. doi: 10.1038/ismej.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng Y, Jiang Y-H, Yang Y, He Z, Luo F, Zhou J. 2012. Molecular ecological network analyses. BMC Bioinformatics 13:113–120. doi: 10.1186/1471-2105-13-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Zhang R, Zheng Q, Deng Y, Van Nostrand JD, Zhou J, Jiao N. 2016. Bacterioplankton community resilience to ocean acidification: evidence from microbial network analysis. ICES J Mar Sci 73:865–875. doi: 10.1093/icesjms/fsv187. [DOI] [Google Scholar]
- 19.de Vries FT, Griffiths RI, Bailey M, Craig H, Girlanda M, Gweon HS, Hallin S, Kaisermann A, Keith AM, Kretzschmar M, Lemanceau P, Lumini E, Mason KE, Oliver A, Ostle N, Prosser JI, Thion C, Thomson B, Bardgett RD. 2018. Soil bacterial networks are less stable under drought than fungal networks. Nat Commun 9:3033–3012. doi: 10.1038/s41467-018-05516-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo J, Guo X, Liang J, Song Y, Liu Y, Li J, Du Y, Mu Q, Jiang Y, Zhao H, Li T. 2021. The influence of elevated CO2 on bacterial community structure and its co-occurrence network in soils polluted with Cr2O3 nanoparticles. Sci Total Environ 779:146430. doi: 10.1016/j.scitotenv.2021.146430. [DOI] [PubMed] [Google Scholar]
- 21.Mendes LW, Raaijmakers JM, de Hollander M, Mendes R, Tsai SM. 2018. Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. ISME J 12:212–224. doi: 10.1038/ismej.2017.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gunawan C, Marquis CP, Amal R, Sotiriou GA, Rice SA, Harry EJ. 2017. Widespread and indiscriminate nanosilver use: genuine potential for microbial resistance. ACS Nano 11:3438–3445. doi: 10.1021/acsnano.7b01166. [DOI] [PubMed] [Google Scholar]
- 23.Grillo R, Rosa AH, Fraceto LF. 2015. Engineered nanoparticles and organic matter: a review of the state-of-the-art. Chemosphere 119:608–619. doi: 10.1016/j.chemosphere.2014.07.049. [DOI] [PubMed] [Google Scholar]
- 24.Khosravi-Katuli K, Shabani A, Paknejad H, Imanpoor MR. 2018. Comparative toxicity of silver nanoparticle and ionic silver in juvenile common carp (Cyprinus carpio): accumulation, physiology and histopathology. J Hazard Mater 359:373–381. doi: 10.1016/j.jhazmat.2018.07.064. [DOI] [PubMed] [Google Scholar]
- 25.Gambardella C, Costa E, Piazza V, Fabbrocini A, Magi E, Faimali M, Garaventa F. 2015. Effect of silver nanoparticles on marine organisms belonging to different trophic levels. Mar Environ Res 111:41–49. doi: 10.1016/j.marenvres.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Bao S, Tang W, Fang T. 2020. Sex-dependent and organ-specific toxicity of silver nanoparticles in livers and intestines of adult zebrafish. Chemosphere 249:126172. doi: 10.1016/j.chemosphere.2020.126172. [DOI] [PubMed] [Google Scholar]
- 27.Westerfield M. 1995. The zebrafish book: a guide for the laboratory use of zebrafish (Brachydanio rerio). University of Oregon Press, Eugene, OR. [Google Scholar]
- 28.Hill AJ, Teraoka H, Heideman W, Peterson RE. 2005. Zebrafish as a model vertebrate for investigating chemical toxicity. Toxicol Sci 86:6–19. doi: 10.1093/toxsci/kfi110. [DOI] [PubMed] [Google Scholar]
- 29.Rawls JF, Samuel BS, Gordon JI. 2004. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci USA 101:4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao F, Zhu W, Yu Y, He Z, Wu B, Wang C, Shu L, Li X, Yin H, Wang J, Juneau F, Zheng X, Wu Y, Li J, Chen X, Hou D, Huang Z, He J, Xu G, Xie L, Huang J, Yan Q. 2021. Host development overwhelms environmental dispersal in governing the ecological succession of zebrafish gut microbiota. NPJ Biofilms Microbiomes 7:5. doi: 10.1038/s41522-020-00176-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan Q, van der Gast CJ, Yu Y. 2012. Bacterial community assembly and turnover within the intestines of developing zebrafish. PLoS One 7:e30603. doi: 10.1371/journal.pone.0030603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis DJ, Bryda EC, Gillespie CH, Ericsson AC. 2016. Microbial modulation of behavior and stress responses in zebrafish larvae. Behav Brain Res 311:219–227. doi: 10.1016/j.bbr.2016.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeLong EF. 2014. Alien invasions and gut “island biogeography. Cell 159:233–235. doi: 10.1016/j.cell.2014.09.043. [DOI] [PubMed] [Google Scholar]
- 34.Yan Q, Li J, Yu Y, Wang J, He Z, Van Nostrand JD, Kempher ML, Wu L, Wang Y, Liao L, Li X, Wu S, Ni J, Wang C, Zhou J. 2016. Environmental filtering decreases with fish development for the assembly of gut microbiota. Environ Microbiol 18:4739–4754. doi: 10.1111/1462-2920.13365. [DOI] [PubMed] [Google Scholar]
- 35.Wu Y, Xiao F, Wang C, Shu L, Zheng X, Xu K, Yu X, Zhang K, Luo H, Yang Y, He Z, Yan Q. 2020. The beta-diversity of Siganus fuscescens-associated microbial communities from different habitats increases with body weight. Front Microbiol 11:1562. doi: 10.3389/fmicb.2020.01562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Umu ÖC, Frank JA, Fangel JU, Oostindjer M, da Silva CS, Bolhuis EJ, Bosch G, Willats WG, Pope PB, Diep DB. 2015. Resistant starch diet induces change in the swine microbiome and a predominance of beneficial bacterial populations. Microbiome 3:16–15. doi: 10.1186/s40168-015-0078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P. 2017. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol 15:630–638. doi: 10.1038/nrmicro.2017.58. [DOI] [PubMed] [Google Scholar]
- 38.Shade A, Peter H, Allison SD, Baho DL, Berga M, Bürgmann H, Huber DH, Langenheder S, Lennon JT, Martiny JBH, Matulich KL, Schmidt TM, Handelsman J. 2012. Fundamentals of microbial community resistance and resilience. Front Microbiol 3:417. doi: 10.3389/fmicb.2012.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narrowe AB, Albuthi-Lantz M, Smith EP, Bower KJ, Roane TM, Vajda AM, Miller CS. 2015. Perturbation and restoration of the fathead minnow gut microbiome after low-level triclosan exposure. Microbiome 3:6–18. doi: 10.1186/s40168-015-0069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Auguste M, Lasa A, Pallavicini A, Gualdi S, Vezzulli L, Canesi L. 2019. Exposure to TiO2 nanoparticles induces shifts in the microbiota composition of Mytilus galloprovincialis hemolymph. Sci Total Environ 670:129–137. doi: 10.1016/j.scitotenv.2019.03.133. [DOI] [PubMed] [Google Scholar]
- 41.Galand PE, Lucas S, Fagervold SK, Peru E, Pruski AM, Vétion G, Dupuy C, Guizien K. 2016. Disturbance increases microbial community diversity and production in marine sediments. Front Microbiol 7:1950. doi: 10.3389/fmicb.2016.01950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Violle C, Pu Z, Jiang L. 2010. Experimental demonstration of the importance of competition under disturbance. Proc Natl Acad Sci USA 107:12925–12929. doi: 10.1073/pnas.1000699107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dethlefsen L, Relman DA. 2011. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA 108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shin N-R, Whon TW, Bae J-W. 2015. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol 33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 45.Zhu D, Zheng F, Chen Q-L, Yang X-R, Christie P, Ke X, Zhu Y-G. 2018. Exposure of a soil collembolan to Ag nanoparticles and AgNO3 disturbs its associated microbiota and lowers the incidence of antibiotic resistance genes in the gut. Environ Sci Technol 52:12748–12756. doi: 10.1021/acs.est.8b02825. [DOI] [PubMed] [Google Scholar]
- 46.Patsiou D, del Rio-Cubilledo C, Catarino AI, Summers S, Fahmi AM, Boyle D, Fernandes TF, Henry TB. 2020. Exposure to Pb-halide perovskite nanoparticles can deliver bioavailable Pb but does not alter endogenous gut microbiota in zebrafish. Sci Total Environ 715:136941. doi: 10.1016/j.scitotenv.2020.136941. [DOI] [PubMed] [Google Scholar]
- 47.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roeselers G, Mittge EK, Stephens WZ, Parichy DM, Cavanaugh CM, Guillemin K, Rawls JF. 2011. Evidence for a core gut microbiota in the zebrafish. ISME J 5:1595–1608. doi: 10.1038/ismej.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collins JW, Keeney KM, Crepin VF, Rathinam VA, Fitzgerald KA, Finlay BB, Frankel G. 2014. Citrobacter rodentium: infection, inflammation and the microbiota. Nat Rev Microbiol 12:612–623. doi: 10.1038/nrmicro3315. [DOI] [PubMed] [Google Scholar]
- 50.Chase JM, Kraft NJ, Smith KG, Vellend M, Inouye BD. 2011. Using null models to disentangle variation in community dissimilarity from variation in α-diversity. Ecosphere 2:art24. doi: 10.1890/ES10-00117.1. [DOI] [Google Scholar]
- 51.Dini-Andreote F, Stegen JC, Van Elsas JD, Salles JF. 2015. Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc Natl Acad Sci USA 112:E1326–E1332. doi: 10.1073/pnas.1414261112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vellend M. 2010. Conceptual synthesis in community ecology. Q Rev Biol 85:183–206. doi: 10.1086/652373. [DOI] [PubMed] [Google Scholar]
- 53.Yan Q, Stegen JC, Yu Y, Deng Y, Li X, Wu S, Dai L, Zhang X, Li J, Wang C, Ni J, Li X, Hu H, Xiao F, Feng W, Ning D, He Z, Van Nostrand JD, Wu L, Zhou J. 2017. Nearly a decade-long repeatable seasonal diversity patterns of bacterioplankton communities in the eutrophic Lake Donghu (Wuhan, China). Mol Ecol 26:3839–3850. doi: 10.1111/mec.14151. [DOI] [PubMed] [Google Scholar]
- 54.Hunt DE, Ward CS. 2015. A network-based approach to disturbance transmission through microbial interactions. Front Microbiol 6:1182. doi: 10.3389/fmicb.2015.01182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Faust K, Raes J. 2012. Microbial interactions: from networks to models. Nat Rev Microbiol 10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 56.Gupta S, Fernandes J, Kiron V. 2019. Antibiotic-induced perturbations are manifested in the dominant intestinal bacterial phyla of Atlantic salmon. Microorganisms 7:233. doi: 10.3390/microorganisms7080233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dai W, Chen J, Xiong J. 2019. Concept of microbial gatekeepers: Positive guys? Appl Microbiol Biotechnol 103:633–641. doi: 10.1007/s00253-018-9522-3. [DOI] [PubMed] [Google Scholar]
- 58.Ayres JS. 2016. Cooperative microbial tolerance behaviors in host-microbiota mutualism. Cell 165:1323–1331. doi: 10.1016/j.cell.2016.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thébault E, Fontaine C. 2010. Stability of ecological communities and the architecture of mutualistic and trophic networks. Science 329:853–856. doi: 10.1126/science.1188321. [DOI] [PubMed] [Google Scholar]
- 60.Marcos MS, Bertiller MB, Olivera NL. 2019. Microbial community composition and network analyses in arid soils of the Patagonian Monte under grazing disturbance reveal an important response of the community to soil particle size. Appl Soil Ecol 138:223–232. doi: 10.1016/j.apsoil.2019.03.001. [DOI] [Google Scholar]
- 61.Dai W, Sheng Z, Chen J, Xiong J. 2020. Shrimp disease progression increases the gut bacterial network complexity and abundances of keystone taxa. Aquaculture 517:734802. doi: 10.1016/j.aquaculture.2019.734802. [DOI] [Google Scholar]
- 62.Ray A, Ghosh K, Ringø E. 2012. Enzyme-producing bacteria isolated from fish gut: a review. Aquacult Nutr 18:465–492. doi: 10.1111/j.1365-2095.2012.00943.x. [DOI] [Google Scholar]
- 63.Ma J, Chen Q-L, O’Connor P, Sheng GD. 2020. Does soil CuO nanoparticles pollution alter the gut microbiota and resistome of Enchytraeus crypticus? Environ Pollut 256:113463. doi: 10.1016/j.envpol.2019.113463. [DOI] [PubMed] [Google Scholar]
- 64.Fuerst JA, Sagulenko E. 2011. Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat Rev Microbiol 9:403–413. doi: 10.1038/nrmicro2578. [DOI] [PubMed] [Google Scholar]
- 65.Feng K, Zhang Z, Cai W, Liu W, Xu M, Yin H, Wang A, He Z, Deng Y. 2017. Biodiversity and species competition regulate the resilience of microbial biofilm community. Mol Ecol 26:6170–6182. doi: 10.1111/mec.14356. [DOI] [PubMed] [Google Scholar]
- 66.Magoč T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stegen JC, Lin X, Fredrickson JK, Chen X, Kennedy DW, Murray CJ, Rockhold ML, Konopka A. 2013. Quantifying community assembly processes and identifying features that impose them. ISME J 7:2069–2079. doi: 10.1038/ismej.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou J, Deng Y, Luo F, He Z, Tu Q, Zhi X. 2010. Functional molecular ecological networks. mBio 1:e00169-10. doi: 10.1128/mBio.00169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’hara R, Simpson GL, Solymos P, Stevens MHH, Wagner H. 2013. Package ‘vegan’. Community ecology package, version 2. http://CRAN.R-project.org/package=vegan. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative abundance of dominant phyla of zebrafish gut microbiota at 15 days of exposure. Different letters above the bars indicate significant differences among groups (ANOVA, P < 0.05), whereas the same letter suggests no significant difference. Download FIG S1, TIF file, 0.2 MB (156.4KB, tif) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Core OTUs (shared by more than 80% of the samples and with a relative abundance of >1%) for different treatments. The relative abundances (A) and a Venn diagram showing the number of shared and unique core OTUs (B) in different treatments are shown. Download FIG S2, TIF file, 1.0 MB (1MB, tif) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Taxonomy of each core OTU (shared by >80% samples and with abundances of >1%) at two sampling times (45 days and 75 days). Download Table S1, DOCX file, 0.02 MB (20.1KB, docx) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A and B) Correlation-based network of zebrafish gut microbial OTUs detected in groups at 45 days (A) and 75 days (B) of exposure. Each node corresponds to an OTU. (C) Zi-Pi plots show the distribution of OTUs based on their topological roles in two exposure groups. (D) Abundance of keystone OTUs. Threshold values of Zi and Pi for categorizing OTUs were 2.5 and 0.62, respectively. Download FIG S3, TIF file, 2.3 MB (2.3MB, tif) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Highly connected modules within zebrafish gut microbial networks for different treatments (control, exposure, and recovery). Colors of nodes indicate different major phyla. Red links indicate positive correlations between two individual nodes, whereas blue links indicate negative correlations between nodes. Download FIG S4, TIF file, 2.9 MB (2.9MB, tif) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Characteristics of AgNPs. (A) AgNP image taken by a transmission electron microscope (TEM). (B) Particle size dispersion of the AgNPs measured by dynamic light scattering (DLS). (C) X-ray diffraction (XRD) patterns of AgNPs. FIG S5, TIF file, 1.4 MB (1.4MB, tif)
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The raw sequencing data are available at the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) under accession number PRJNA668536.