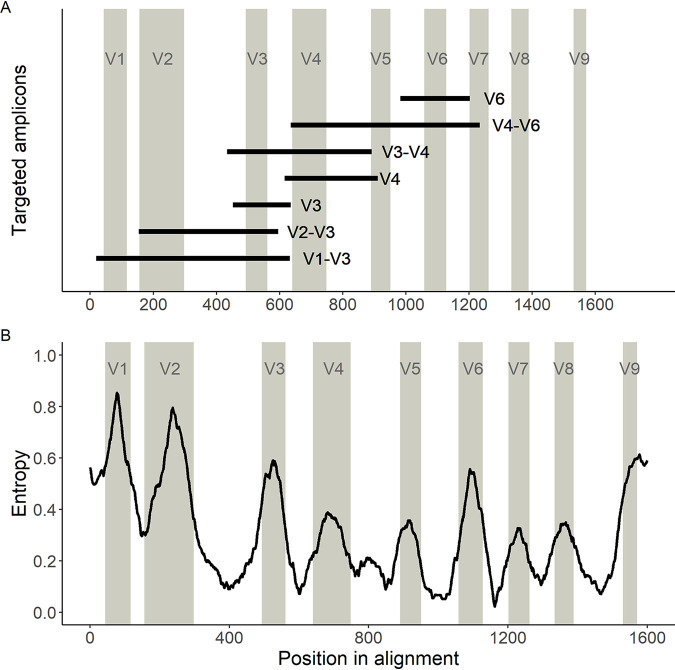
FIG 2.
Variable regions of the 16S rRNA gene used in this study. (A) Locations of the primers used in this study on the 16S rRNA gene. Locations of the predicted amplicons are shown as black bars in relation to the multiple-sequence alignment (MSA) of the bacterial species described in Thomas-White et al. (33). Gray columns are the locations of the known variable regions based on the sequence from Escherichia coli. (B) Information about variable regions, measured by entropy from a sliding-window analysis of the MSA. Higher entropy indicates that the region has more variability across species and therefore more information to identify a bacterial species. Lower entropy indicates that the region has little variability (i.e., is conserved) across species and therefore less information to identify a bacterial species.