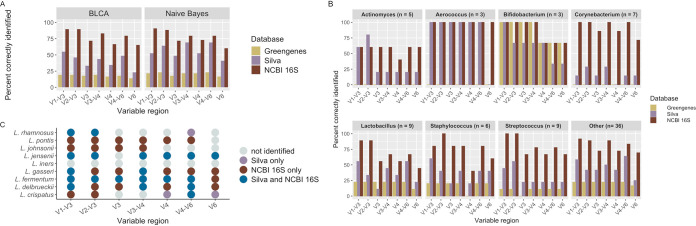
FIG 5.
Percentage of bladder bacteria correctly identified for each classification scheme. (A) Overall results by classifier. With the commonly used V4 variable region and BLCA classifier, 17% of bladder bacteria are correctly identified using the Greengenes database, compared with 35% correctly identified using the Silva database and 67% using the NCBI 16S database. A similar trend is seen with the naive Bayes classifier. Using other variable regions can lead to improved species-level resolution to a maximum of 91% correctly identified. (B) Percentage of bladder bacterial species identified with each database by variable region (BLCA). Genera with fewer than 3 species are grouped as “other.” (C) Identification of Lactobacillus species depends on the variable region and database used, with the NCBI 16S database having the most coverage, regardless of the variable region chosen. The Greengenes database is not shown, since it only classified two species (Lactobacillus pontis and Lactobacillus delbrueckii).