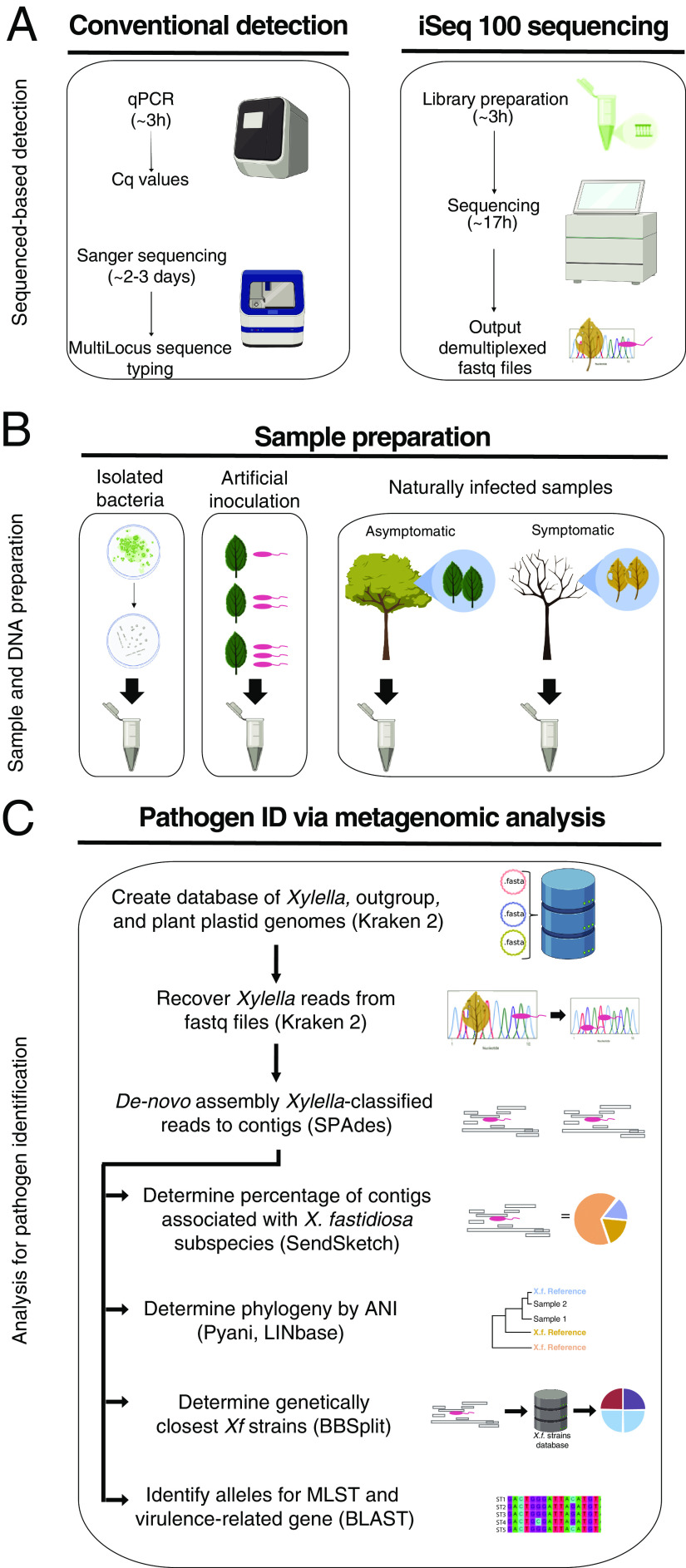
FIG 1.
Metagenomics for the diagnostic pipeline. (A) Sequenced-based detection. Two approaches were used for Xf detection—conventional detection and iSeq 100 sequencing. For conventional detection, samples were analyzed using qPCR assays, Harper’s test or tetraplex Dupas’s test, and MLST involving Sanger sequencing of seven housekeeping genes. iSeq 100 libraries were prepared according to the manufacturer’s instructions. After 17 h of sequencing, demultiplexed samples were recovered from the machine and used for subsequent analysis. (B) Sample preparation. The samples used for the pipeline were DNA-extracted from bacterial strains in culture, spiked plant material, and naturally infected samples. (C) Pathogen identification via metagenomic analysis. Demultiplexed fastq reads from all samples were then used for metagenomic analysis. We created a database to recover Xf reads using Kraken 2. The database contained Xylella, Xanthomonas, and Escherichia coli genomes. We also added plant plastid genomes to remove false-positive results. Xf reads were recovered from the fastq files. The Xf recovered reads were de novo assembled to obtain Xf contigs, using SPAdes. The Xf contigs were used in four different analyses, subspecies identification, phylogeny reconstruction, identification of the genetically closest strain with a sequenced genome, and alleles from specific genes. To determine subspecies, we used the tool SendSketch. To reconstruct phylogeny, we calculated ANI using Pyani and the website tool LINbase (https://linbase.org/). To determine the genetically closest known Xf strain, we detected the number of hits to each Xf strain using BBSplit. To identify specific gene alleles, we calculated the percentage of identity to the seven MLST genes (cysG, gltT, holC, leuA, malF, nuoL, petC) and the percentage of similarity to 17 virulence-related proteins using local BLAST+. Graphics were created with BioRender.