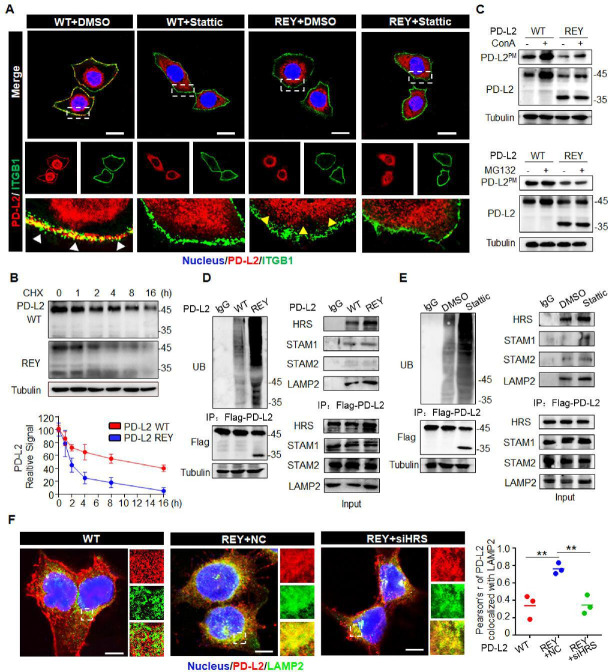
Figure 4.
Glycosylation inhibits ubiquitination-mediated lysosomal degradation of PD-L2 and facilitates PD-L2 membrane abundance. (A) Immunofluorescence staining was performed to examine the expression pattern of PD-L2 in SCC15 cells expressing WT or REY mutant of PD-L2 with or without Stattic (7 µM) treatment for 4 hours. PD-L2 WT was mainly localized to the cell membrane (white arrow), colocalized with ITGB1. Stattic treatment resulted PD-L2 WT localized in the cytosol. PD-L2 REY mutant was primarily localized to the cytoplasm with or without Stattic treatment (yellow arrow). Scale bar, 20 µm. (B) SCC15 cells expressing PD-L2 WT or REY were treated with 20 µM CHX at the indicated time points. Bottom panel shows the quantification of the data. (C) Membrane PD-L2 (PD-L2PM) levels in cells expressing PD-L2 WT or REY treated with the lysosome inhibitor concanamycin-A or proteasome inhibitor MG132. (D) The REY mutant markedly increased PD-L2 ubiquitination when compared with WT PD-L2. Co-immunoprecipitation (Co-IP) analysis of the interaction between HRS, LAMP2, STAM1, STAM2, and PD-L2 in SCC15 cells expressing PD-L2 WT or REY. (E) Effect of the STAT3 inhibitor Stattic on PD-L2 ubiquitination. Co-IP was performed to analyze the interaction between HRS, LAMP2, STAM1, STAM2, and PD-L2 in the Stattic-treated cells. (F) Colocalization of PD-L2 with the lysosome marker LAMP2 in HRS knockdown SCC15 cells expressing PD-L2 WT or REY. Scale bar, 5 µm. Right panels show the quantification of the colocalization between PD-L2 and LAMP2. Values represent means±SD from three independent experiments. Significance was detected using one-way analysis of variance. **p<0.01. PD-L2, programmed death ligand 2; REY, PD-L2 REY mutant, in which the amino acid of R168, E171 and Y174 were substituted by Alanine; STAT3, signal transducer and activator of transcription 3; WT, wild type.