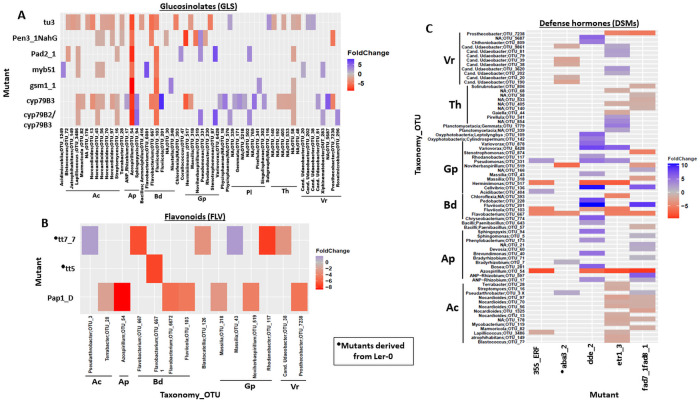
Fig 5. Heat map of differentially abundant bacterial OTUs identified by comparing secondary metabolite and signalling mutants and their parental lines.
A) Heat map of differentially abundant bacterial OTUs identified by comparing GLS mutants and parental lines. B) Heat map of differentially abundant bacterial OTUs identified by comparing FLVs mutants and parental lines. C) Heat map of differentially abundant bacterial OTUs identified by comparing DSMs mutants and parental lines. Foldchanges are indicated in blue and red respectively for parental lines and mutants. ANP-Rhizobium (Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium), Candidatus Udaeobacter (Cand. Udaeobacter), Candidatus Xiphinematobacter (Cand. Xiphinematobacter), Ac (Actinobacteria), Ap (Alphaproteobacteria), Bd (Bacteroidetes), Gp (Gammaproteobacteria), Pl (Planctomycetes), Th (Thermoleophilia), Vr (Verrucomicrobiae). Analysis was performed using DESeq2 package.