Figure 1. Sugars control alternative splicing in roots.
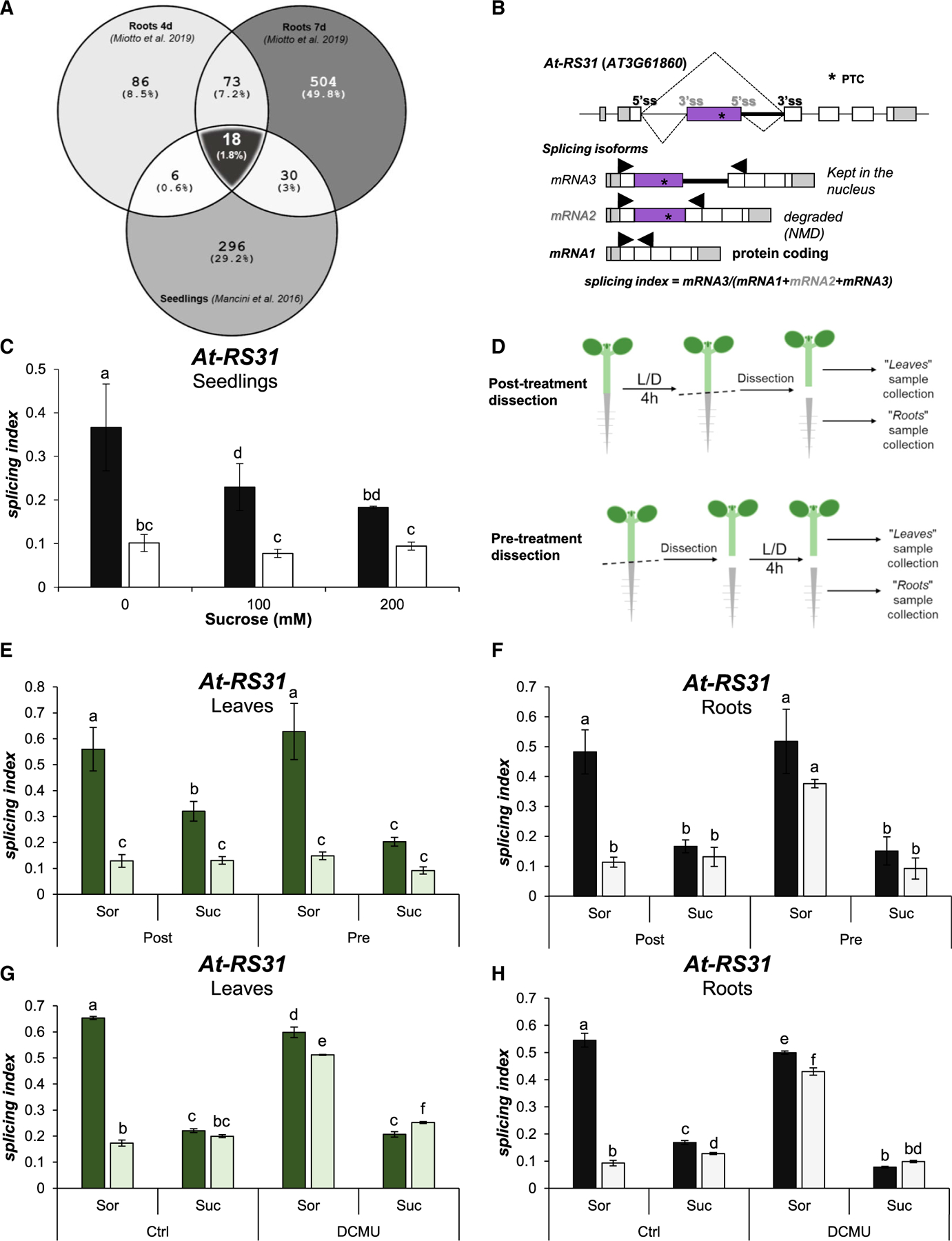
(A) Venn diagram showing overlap between affected alternative splicing events in RNA-seq data from different experiments. Differentially alternatively spliced (DAS) genes were assessed in data from root samples, with shoots exposed to long-day conditions or darkness (Miotto et al., 2019), and data from seedling samples that received an acute light treatment at the middle of the night period (Mancini et al., 2016). Significantly affected genes are listed in Tables S1, S2, and S3.
(B) Gene model and alternative splicing isoforms of At-RS31. *, PTC: premature termination codon. Arrows: primers used for splicing evaluation. The alternative usage of 5′ss and 3′ss gives rise to 3 isoforms. Usage of gray 3′ss generates mRNA3, and if the gray 5′ss is also recognized, mRNA2 (inclusion of purple exon). The coding isoform, mRNA1, is generated by the use of black 5″ and 3′ss only.
(C) Exogenous sucrose (Suc) diminishes splicing indexes in dark-treated seedlings. Whole seedlings were treated under a light/dark protocol (Figure S1) and sucrose was added to plant growth media at 100 or 200 mM. Sorbitol (Sor) was used as osmotic control to ensure equal osmolarity (200 mM total) in all of the treatments.
(D) Post-treatment dissection was done after the light/dark treatment, immediately before sample collection. Pre-treatment dissection was done before light/dark incubation.
(E and F) Roots are not directly responsive to light but they are responsive to sugars. Incubation with sucrose (200 mM) was conducted during the light/dark treatments.
(G and H) Sugar control of alternative splicing is independent of chloroplast function. Sucrose (100 mM) and DCMU (20 μM) were added before light/dark treatments. Ethanol was used as vehicle (Ctrl). In (C) and (E)-(H), Sorbitol (Sor) was used as osmotic control at the same concentration as sucrose. Lighter bars, light; darker bars, darkness. Data represent splicing index means ± standard errors (n = 4). The same letters indicate means that are not statistically different (p > 0.05).
