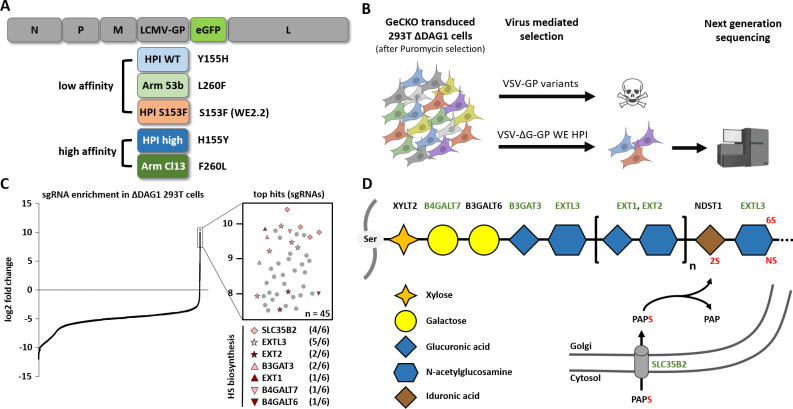
Fig 1. Genome-wide CRISPR Cas9 knockout screen identifies host factors import for LCMV infection in the absence of DAG1.
(A) Schematic overview of the genome of replication competent VSV-based reporter systems pseudotyped with high or low affinity LCMV GP variants and eGFP as a transgene. (B) Schematic overview of CRISPR Cas9-mediated knockout screen in 293T ΔDAG1 cells with replication competent or incompetent high and low affinity VSV-GP variants. Created with BioRender.com. (C) Enrichment of sgRNA expression after low affinity VSV-ΔG-GP (HPI WT) selection compared to an untreated control (left). Representation of top hits and number of significantly enriched sgRNAs (out of 6) per gene (right; n = 45) and (D) their roles in the heparan sulfate biosynthesis pathway. Enriched genes from the genome-wide CRISPR Cas9 screen are marked in green.