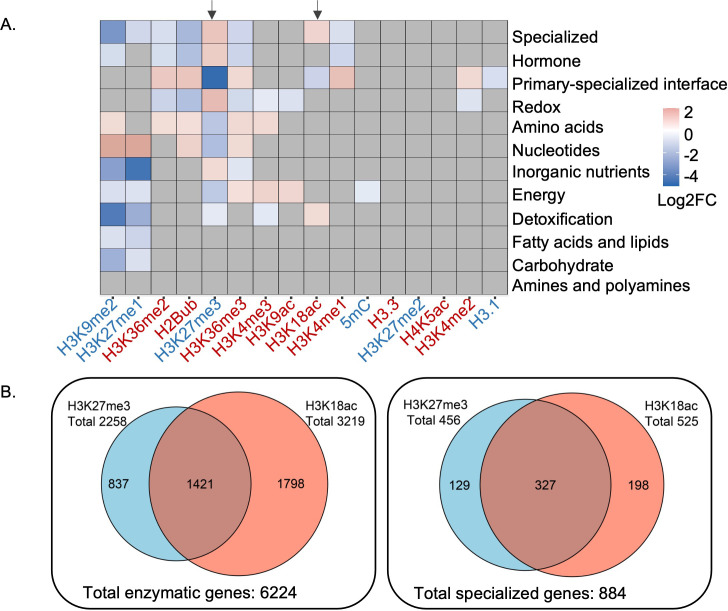
Figure 1. Patterns of epigenetic modification across metabolism in Arabidopsis.
(A) Enrichment analysis shows different epigenetic modification patterns across metabolic domains. The heatmap represents log2 fold change (Log2FC) of enrichment or depletion of an epigenetic mark associated with each domain relative to total metabolic genes. Epigenetic modifications are color-coded based on their effect on gene expression; red represents activating mark and blue represents repressive mark. Genes only mapped to unique metabolic domains were included in this analysis. Significant enrichment or depletion is based on Fisher’s exact test, p-value < 0.05 and fold change >1.5. Gray cells represent no significant change. Black arrows indicate the patterns for trimethylation of lysine 27 of histone 3 (H3K27me3) and H3K18ac. (B) The co-occurrence of H3K27me3 and H3K18ac on total enzymatic genes in the genome and specialized metabolic genes.