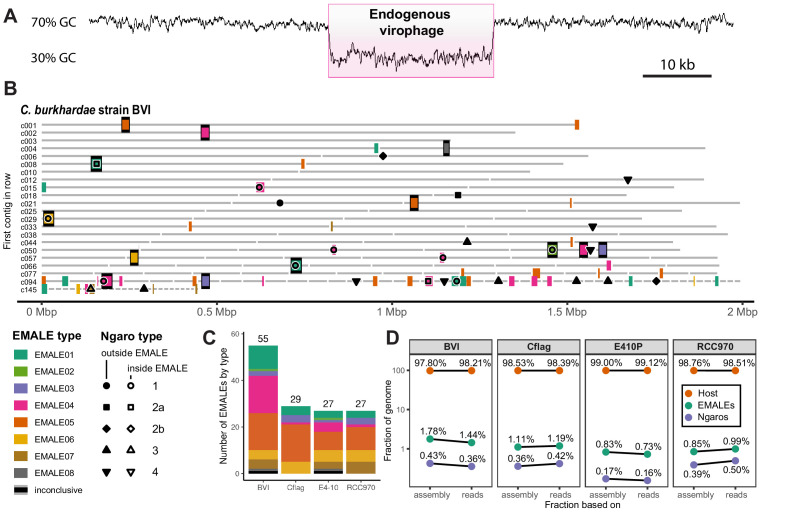
Figure 1. Endogenous virophages in Cafeteria burkhardae.
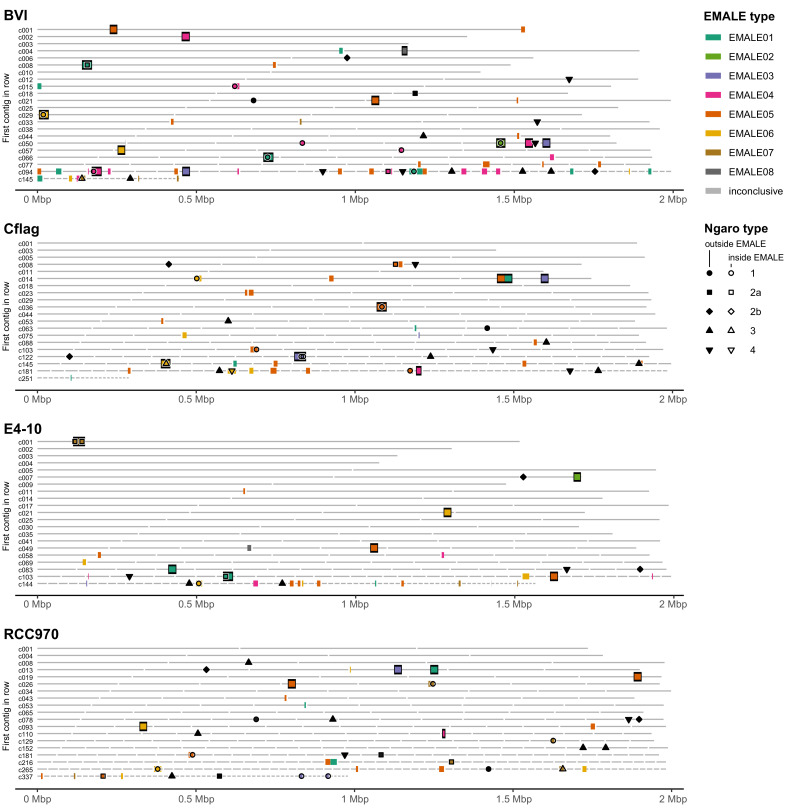
(A) GC-content graph signature of a virophage element embedded in a high-GC host genome. Shown is a region of contig BVI_c002 featuring an integrated virophage (pink box) flanked by host sequences. (B) Location of partial or complete virophage genomes and Ngaro retrotransposons in the genome assemblies of C. burkhardae strain BVI (see Figure 1—figure supplement 1 for all four strains). Horizontal lines represent contigs of decreasing length ordered from left to right and from top to bottom, with numbers shown for the first contig of each line; colored boxes indicate endogenous mavirus-like elements (EMALEs). Fully assembled elements are framed in black. Ngaro retrotransposon positions are marked by black symbols; open symbols indicate Ngaros integrated inside a virophage element. (C) Graphic summary of the number and types of all EMALEs identified in each of the four C. burkhardae strains. (D) Nucleotide contributions of EMALEs and Ngaros to Cafeteria genomes. Fractions for each strain are computed based on nucleotides in the assembly (left) and nucleotides in the reads (right) mapping to the different parts of the assembly.