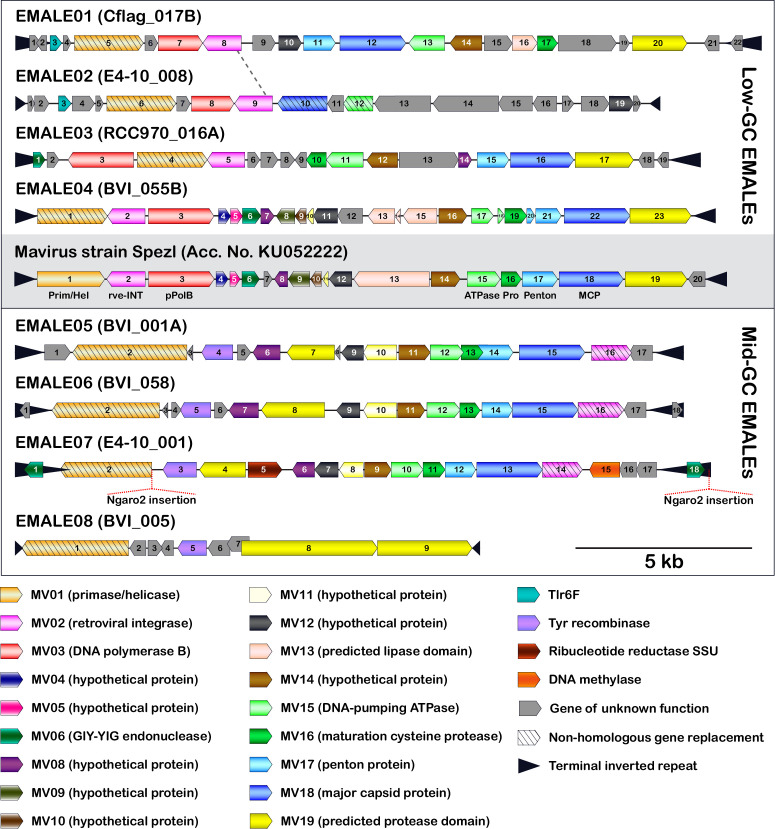
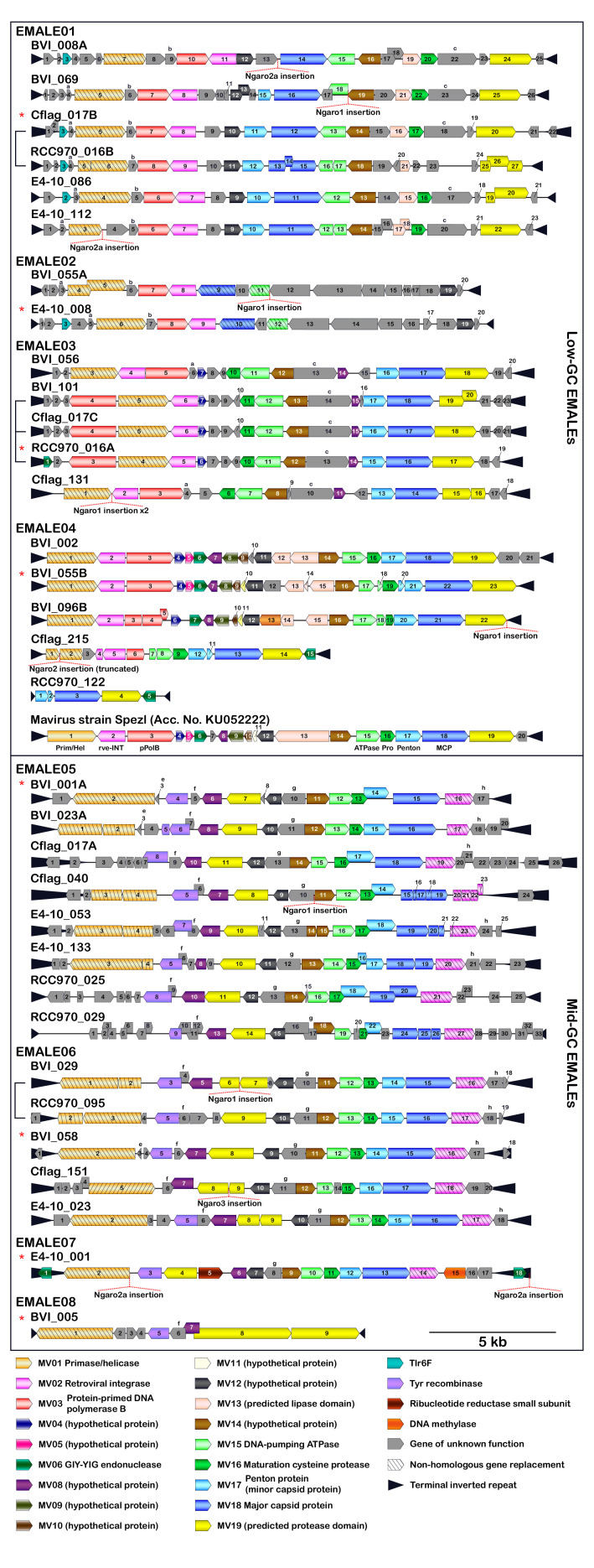
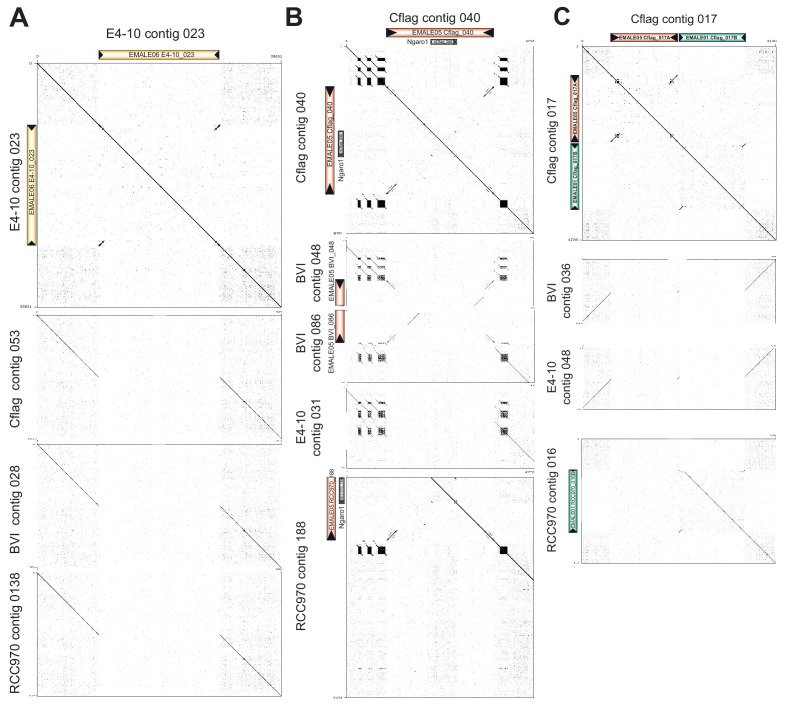
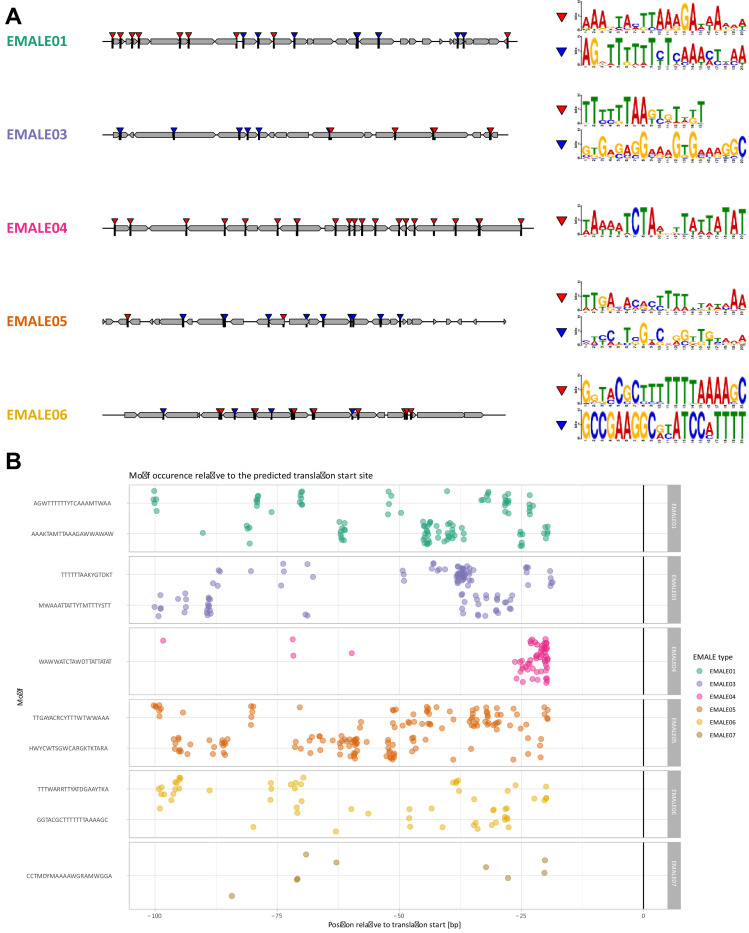
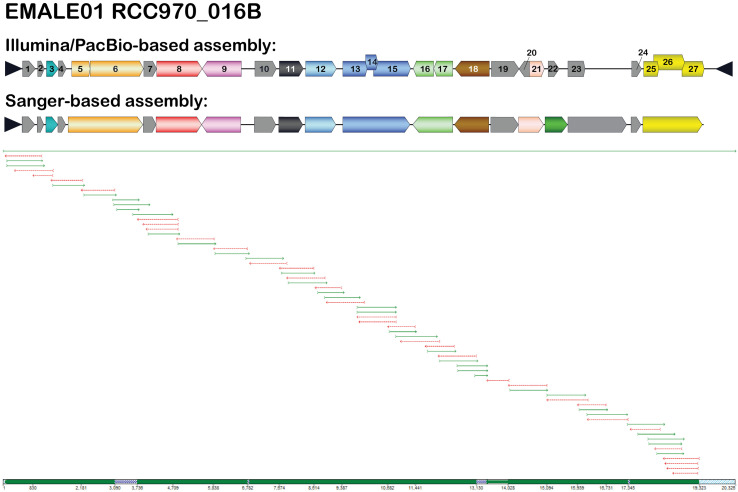
Figure 3. Genome organization of eight EMALE types found in Cafeteria burkhardae.
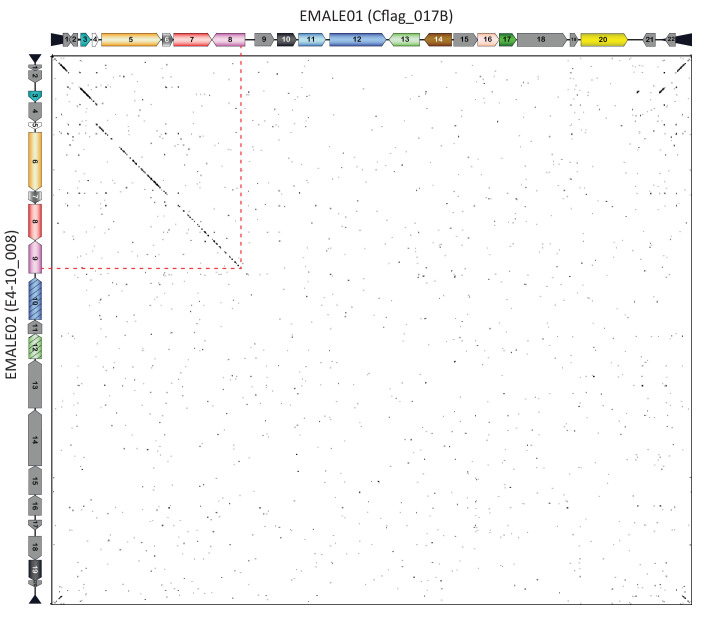
Shown are schematic genome diagrams of the EMALE type species 1–8; for all 33 complete EMALEs, see Figure 3—figure supplement 1. The reference mavirus genome with genes MV01-MV20 is included for comparison. Homologous genes are colored identically; genes sharing functional predictions but lacking sequence similarity to the mavirus homolog are hatched. Open reading frames are numbered individually for each element. Ngaro retrotransposon insertion sites are indicated where present. The dotted line between EMALE01 and EMALE02 separates a homologous region (left) from unrelated DNA sequences (right) and thus indicates the location of a probable recombination event.