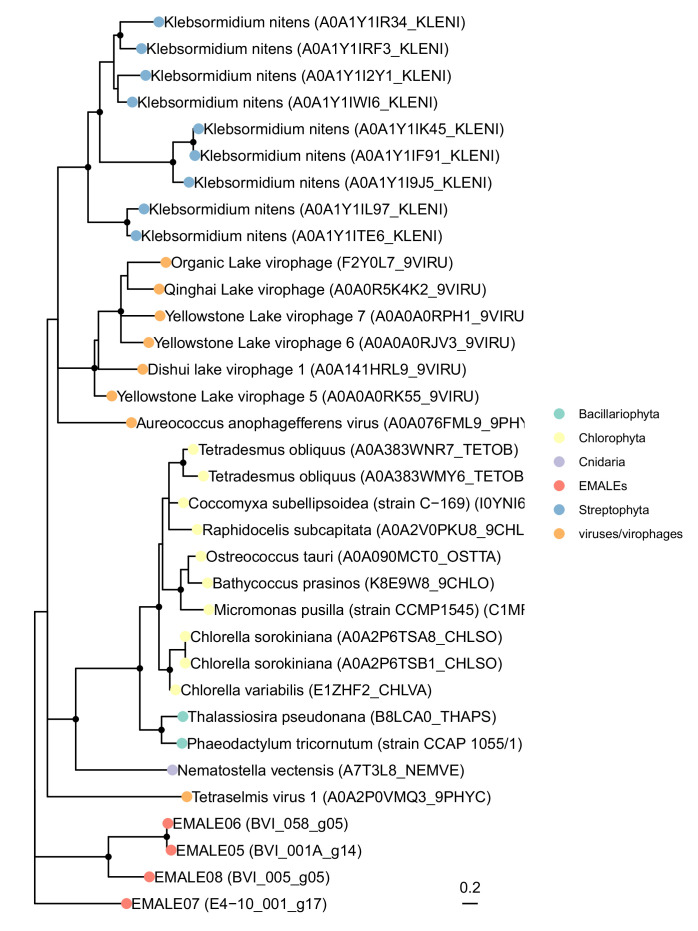
Figure 4. Phylogenetic reconstruction of conserved EMALE proteins.
Unrooted maximum likelihood trees were constructed from multiple sequence alignments of the four virophage core proteins major capsid protein (MCP), penton protein (PEN), ATPase, and protease (PRO), as well as of the retroviral integrase. Nodes with bootstrap values of 80% or higher are marked with dots. EMALEs are color-coded by type; cultured virophages are printed in bold. ALM, Ace Lake Mavirus; DSLV, Dishui Lake virophage; OLV, Organic Lake virophage; RVP, rumen virophage; TBE/TBH, Trout Bog Lake epi-/hypolimnion; YSLV, Yellowstone Lake virophage. Metagenomic sequences starting with Ga and M590 are derived from Paez-Espino et al., 2019.