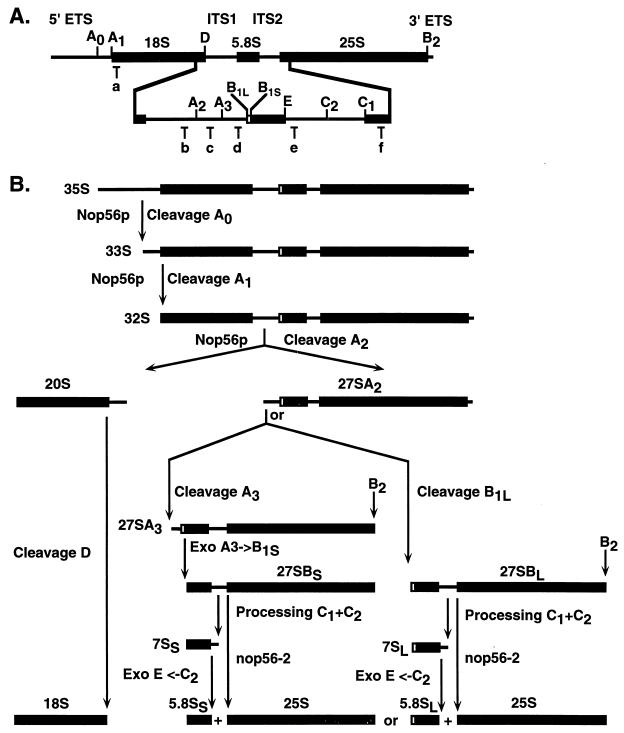
FIG. 1.
Structure of the ribosomal DNA and pre-rRNA-processing pathway in yeast. (A) In the 35S primary transcript, the sequences of the mature 18S, 5.8S, and 25S pre-rRNAs are flanked by the external transcribed spacers (5′ and 3′ ETS) and separated by the internal transcribed spacers (ITS1 and ITS2). The cleavage sites are indicated by uppercase letters (A0 to E), and the oligonucleotide probes used are indicated by lowercase letters (a to f). (B) Successive cleavage of the 35S pre-rRNA at sites A0 and A1 generates the 33S and 32S pre-rRNAs. Cleavage of the 32S pre-rRNA at site A2 then generates the 20S and 27SA2 pre-rRNAs, which are precursors to the RNA components of the small and large ribosomal subunits, respectively. The mature 18S rRNA is generated by cleavage of the 20S pre-rRNA at site D. The 27SA2 precursor is either cleaved at site A3 by RNase MRP, generating the 27SA3 pre-rRNA, or at site B1L to yield 27SBL pre-rRNA. The 27SA3 pre-rRNA is rapidly digested by the 5′-to-3′ exonucleases Xrn1p and Rat1p to yield the 27SBS pre-rRNA. Processing at site B2, the 3′ end of the 25S rRNA, occurs concomitantly with 27SB 5′-end formation, since a probe specific for the 10-nucleotide extension beyond site B2 detects 27SA but not 27SB (21). The 27SBS and 27SBL pre-rRNAs both follow the same pathways of processing to 25S and 5.8SS/L through cleavage at sites C1, the 5′ end of the mature 25S rRNA, and C2 in ITS2, followed by 3′-to-5′ exonucleolytic digestion of 7SS and 7SL from site C2 to E by the exosome complex. Pre-rRNA processing analysis in GAL-regulated and thermosensitive alleles of Nop56p (nop56-2) indicated that Nop56p is required for the early pre-rRNA cleavage at sites A0, A1, and A2. In addition, 27SB processing was inhibited in nop56-2 strains.