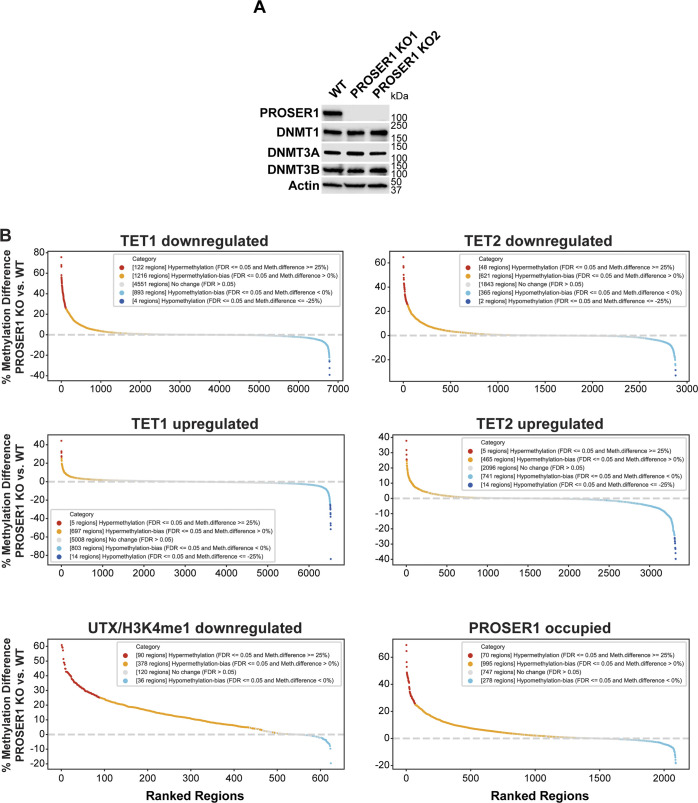
Figure S7. Exploring alternative mechanisms of PROSER1-mediated changes in DNA methylation.
(A) Western blot for members of the DNA methyltransferase family from nuclear extracts of WT and PROSER1 KO HEK293 cells. Actin was used as a loading control. (B) DNA methylation rank plots showing regions associated with TET1 down-regulation, TET2 down-regulation, TET1 up-regulation, TET2 up-regulation, UTX/H3K4me1 down-regulation, and PROSER1-occupied regions in PROSER1 KO versus WT HEK293 cells (all FC ≥ 2). Regions with CpGs covered by at least 10 whole genome bisulfite sequencing reads (please see “whole genome bisulfite sequencing data processing” in the “Materials and Methods” section for further details) were ranked from hyper- to hypomethylated on the x-axis.