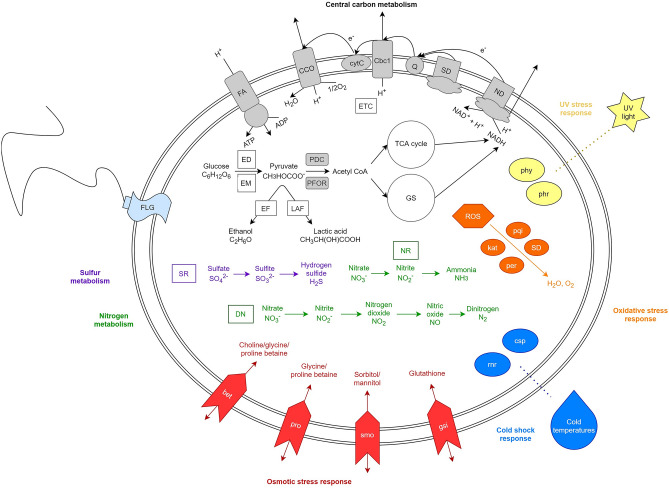
Figure 5.
Model of Hybrid_5 cellular systems based on genomic data. Central carbon, nitrogen, and sulfur metabolisms are present, as well as stress response genes and pathways. Central carbon metabolism: Cbc1 = cytochrome bc1 complex; CCO = cytochrome c oxidase, cbb3-type; cytC = cytochrome C; ED = Entner-Doudoroff pathway; EF = ethanol fermentation; EM = Embden-Meyerhof glycolysis; ETC = electron transport chain; FA = F-type ATPase; Q = ubiquinone/quinone pool; GS = glyoxylate shunt; LAF = lactic acid fermentation; ND = NADH dehydrogenase; PDC = pyruvate dehydrogenase complex; PFOR = pyruvate:ferredoxin oxidoreductase; SD = succinate dehydrogenase; TCA = tricarboxylic acid. Nitrogen metabolism: DN = denitrification; NR = nitrate reduction. Sulfur metabolism: SR = sulfur reduction. Cold shock response: csp = cold shock protein; rnr = cold shock-induced ribonuclease R. Osmotic stress response: bet = glycine/choline/proline betaine transporters; gsi = glutathione transporters; pro = glycine/proline betaine transporters; smo = sorbitol/mannitol transporters. Oxidative stress response: kat = catalase-peroxidase; per = peroxidase; pqi = paraquat-inducible protein A; ROS = reactive oxygen species; SD = superoxide dismutase; UV stress response: phr = photolyase; phy = phytoene synthase. FLG = flagellar biosynthesis genes.