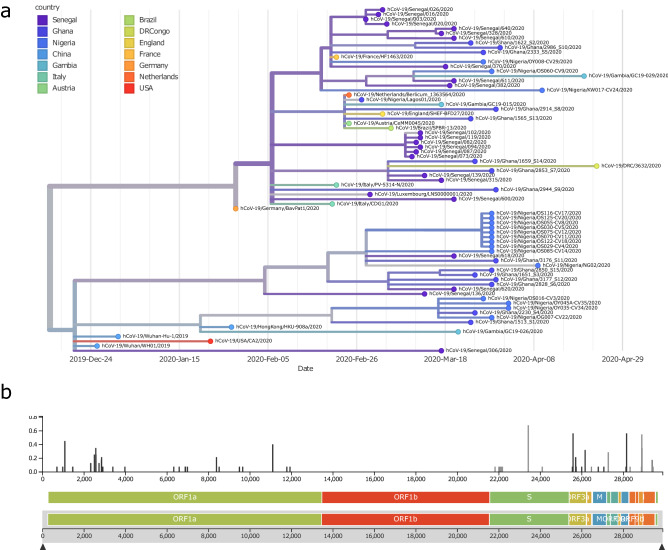
Figure 1.
Phylogenetic tree (December 2019 to April 2020) revealing similarities of West-African viral sequences with Chinese and multiple European countries. (a) Nigerian samples cluster with the early Chinese samples within the bottom branch of the tree, Ghanaian samples are about equally distributed between the top (European) and bottom branch of the tree. Senegalese samples cluster closer with the French reference sample on the top of the tree. (b) Highest diversity is at the A23403G (D614G) substitution splitting the tree in the bottom (Chinese) and top (European) branch. This variant has been reported to increase infectivity1,12. Graphics were generated using the nextstrain pipeline including software Augur (version 7.0.2, https://docs.nextstrain.org/projects/augur/en/stable/index.html) and TreeTime (version 0.7.6, https://github.com/neherlab/treetime)28,31.