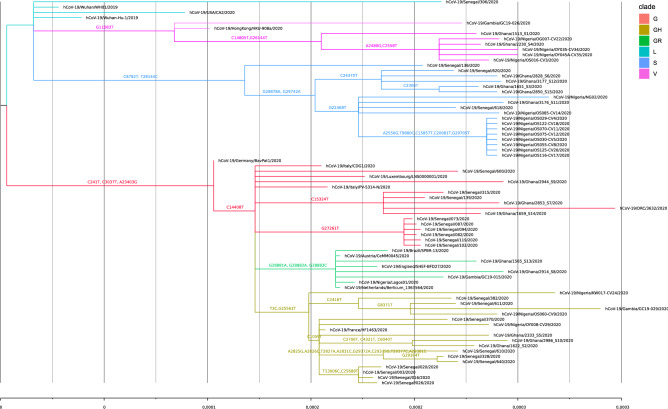
Figure 2.
Phylogenetic tree (December 2019–April 2020) colored by clades shows distribtution of West-African samples over all clades suggesting introductions from China and European countries. Patterns are country-specific, e.g. most Senegalese samples have close similarity with the French reference, most Nigerian samples cluster in early Chinese-based clade S and Ghanaian samples are spread over all clades. Within the clade S, there are putatively specific West-African mutations at the branches at C24370T and G22468T. G22486T may reflect migration routes because in the nextstrain analysis of Africa as a whole there are also Tunisian samples in this branch (https://nextstrain.org/ncov/africa?f_region=Africa, accessed Jun 26th, 2020). Two of the non-French related Senegalese samples come from these branches while the other (Senegal/136) has strong similarity with Spanish-end of February samples from the early clade S pointing at multiple introductions to Senegal from France, Spain and African countries. Graphics were generated using the software FigTree (version 1.4.4, http://tree.bio.ed.ac.uk/software/figtree/) for annotation of the phylogenetic tree and the nextstrain pipeline including software Augur (version 7.0.2, https://docs.nextstrain.org/projects/augur/en/stable/index.html) and TreeTime (version 0.7.6, https://github.com/neherlab/treetime)28,31.