Abstract
A complete understanding of the behavioral influence and phenotypic transition of vascular smooth muscle cells, as well as the effects of the characteristics of these cells on the physiological and pathological processes of atherosclerosis, is crucial if new therapeutic targets for atherosclerosis are to be identified. In the present study, the long non-coding RNA RP11-531A24.3 was identified to be expressed at low levels in plaque tissues through screening a microarray for differentially expressed genes. The functional experimental results suggested that RP11-531A24.3 reduced the viability and inhibited the migration of human aortic vascular smooth muscle cells (HA-VSMCs). RNA antisense purification-mass spectrometry was used to identify the RNA-binding proteins (RBPs) for RP11-531A24.3. Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis indicated that the pathway with the highest degree of association with RP11-531A24.3 RBPs was related to cell migration. The reduced migration and viability mediated by RP11-531A24.3 overexpression was more significantly suppressed after annexin 2 (ANXA2) depletion in RP11-531A24.3-overexpressing HA-VSMCs. Culture of HA-VSMCs under hypoxic conditions (1% O2) reduced the expression of RP11-531A24.3, and enhanced the protein expression of ANXA2 and HIF-1α, while knockdown of ANXA2 downregulated the protein expression of HIF-1α. These results suggested that RP11-531A24.3 regulated the proliferation and migration of HA-VSMCs through ANXA2 expression, and hypoxia may be an external factor in the regulation of RP11-531A24.3 and its downstream targets.
Keywords: vascular smooth muscle cell, proliferation, migration, long non-coding RNA, hypoxia
Introduction
Cardiovascular disease currently ranks as the most common cause of illness and death worldwide, and atherosclerosis is the main underlying pathogenetic mechanism of cardiovascular diseases (1,2). Atherosclerosis is a pathological disease characterized by lipid accumulation, inflammation, fibroproliferation and formation of plaques with a necrotic core that contains a variety of vessel wall cell types (3). Vascular smooth muscle cells (VSMCs) are the primary components of the plaque from which extracellular matrix (ECM) components are derived at all stages of atherosclerosis (4,5). At the pre-atherosclerosis stage, VSMCs in diffuse intimal thickening exhibit reduced expression of contractile proteins and switch to a synthetic phenotype, which leads to increased generation of ECM components rich in proteoglycans; changes in ECM components lead to the retention of apolipoproteins and recruitment of monocytes, triggering inflammation (6,7). Progression from pre- to early atherosclerosis is facilitated by a complex interaction between lipid retention and oxidation, induction of inflammation and proliferation of VSMCs, phenotypic transformation of VSMCs and VSMC death (5,7). In the advanced plaque milieu, where insufficient efferocytosis has occurred, the self-perpetuating inflammatory response leads to an alteration in VSMC phenotypes and changes in cell behavior, including apoptosis, senescence, migration and proliferation; it is therefore critical to identify the environmental cues that regulate phenotypic transitions of VSMCs and the intrinsic mechanisms underlying these (4,5,8).
The anoxemia hypothesis suggests that a mismatch between oxygen supply and demand in the arterial wall leads to the development of lesions and plaques (9-11). Hypoxia-inducible factors (HIFs) are a family of key regulator proteins that are expressed in the hypoxic state, including HIF-1β (whose activity is not affected by hypoxia) and HIF-1α (active subunit with a half-life of 5 min and highly regulated by oxygen). By activating the genes that alter energy metabolism, cell proliferation, angiogenesis and vascular remodeling, HIF-1α allows cells to adapt to a hypoxic environment (10). Hypoxia and particularly HIF-1α expression is crucial for stimulating vascular remodeling through the proliferation and migration of VSMCs (10).
Long non-coding RNAs (lncRNAs) represent a family of non-protein coding transcripts of ~200 nucleotides in length (12-15). lncRNAs regulate gene expression through complex molecular mechanisms at all levels, including the post-transcriptional, transcriptional and chromatin levels, either in the nucleus or in the cytoplasm (15-17). As lncRNAs have high tissue specificity, they exist as vital epigenetic regulators in almost every cellular process in which gene expression is distinctly mediated under physiological and pathological conditions (13,14,16). It has been demonstrated that lncRNAs are involved in plasticity and vascular dysfunction of VSMCs, and directly affect the progression and development of atherosclerosis (16,17).
The emerging links between lncRNAs and atherosclerosis have provided a new perspective to explore the pathogenesis of atherosclerosis, and a new field of opportunities to develop therapeutic and diagnostic approaches (16,18,19). In the present study, the role of lncRNA RP11-531A24.3 in the phenotypic regulation of VSMCs was identified, with a preliminary investigation into its regulation mechanism conducted. Hypoxic culture conditions were also used to evaluate the relationship between RP11-531A24.3 and HIF-1α expression.
Materials and methods
Screening of differentially expressed genes and bioinformatics prediction
By performing microarray analysis with an Agilent-045997 Arraystar human lncRNA microarray V3 (Probe Name Version; Agilent Technologies, Inc.), the lncRNA profile of three advanced atherosclerosis samples and three normal intima tissues was characterized (NCBI accession no. GSE97210; https://www.ncbi.nlm.nih.gov/geo/) (20,21). Prediction of the non-coding sequence region of RP11-531A24.3 was performed using the ORF prediction tools Coding Potential Calculator (http://cpc2.gao-lab.org/) and Coding Potential Assessing Tool (http://lilab.research.bcm.edu/cpat/index.php). The Database for Annotation, Visualization and Integrated Discovery bioinformatics resources, version 6.8 (https://david.ncifcrf.gov/tools.jsp) was used for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (22) and Gene Ontology (GO) (23) enrichment analysis of the RNA-binding proteins of RP11-531A24.3 with P<0.05 and gene count ≥1.
Reagents
Human aortic (HA-)VSMCs (cat. no. CRL-1999) were obtained from the American Type Culture Collection and authenticated via STR profiling. Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), penicillin, streptomycin, puromycin, bovine serum albumin and polybrene were purchased from Gibco (Thermo Fisher Scientific, Inc.). Empty lentivirus vector (anti-puromycin; LV-mock), LV-mediated lncRNA RP11-531A24.3 overexpression vector (anti-puromycin; LV-OE), small interfering (si)RNA targeting annexin A2 (ANXA2) and its corresponding si-negative control (si-NC; cat. no. SIGS0005567-1), and the siRNA targeting HIF-1α and its corresponding si-NC (cat. no. SIGS0002003-1) were constructed and purchased from Guangzhou RiboBio Co., Ltd. An Annexin V-APC/7-aminoactinomyosin (7-AAD) Apoptosis Detection kit (cat. no. KGA1023) and a Cell Cycle Detection kit (cat. no. KGA512) were obtained from Nanjing KeyGen Biotech. Co., Ltd. Cell Counting Kit-8 (CCK-8; cat. no. C0039) was purchased from Beyotime Institute of Biotechnology and Transwell chambers (cat. no. 3422) from BD Biosciences. Lipofectamine 3000 was purchased from Life Technologies. Co., Ltd. TRIzol® was purchased from Invitrogen; Thermo Fisher Scientific, Inc. A PrimeScript II 1st Strand cDNA Synthesis kit and a SYBR Green PCR kit were purchased from Takara Biotechnology Co., Ltd. An All-in-One First-Strand cDNA Synthesis kit was purchased from GeneCopoeia, Inc. RIPA lysis buffer was purchased from Hangzhou Fude Biological Technology Co., Ltd. Antibodies targeting ANXA2 (rabbit polyclonal; cat. no. ab235939) and β-tubulin (rabbit polyclonal; cat. no. ab6046) were purchased from Abcam, while antibody targeting HIF-1α (rabbit polyclonal; cat. no. AF1009) was purchased from Affinity Biosciences. ECL Western Blotting Substrate was purchased from Promega Corporation. RNA Antisense Purification Kit (cat. no. Bes 5103-2; BersinBio) was used in the RNA antisense purification (RAP) experiment (http://bersinbio.com/Product/detail/id/14.html). All reagents were used according to the manufacturers' protocol.
Cell culture
HA-VSMCs were incubated in DMEM containing 100 U/ml penicillin, 10% FBS and 100 mg/ml streptomycin in a humidified atmosphere of 5% CO2 and 95% air at 37˚C. For standard culture, the cells were exposed to co-mixture gas (75% N2, 5% CO2 and 20% O2). For hypoxic culture, the cells were exposed to a co-mixture of hypoxia-inducing gases (94% N2, 5% CO2 and 1% O2) in an impenetrable sealed modular incubator chamber (Shel Lab Bactron series; Sheldon Manufacturing) at 37˚C for 6, 12, and 24 h. Before hypoxic culture, cells had been transfected with si-RNA targeting ANXA2 or HIF-1α.
Tissue samples
Plaque tissues and endothelial tissues (n=5 of each) of suitable size were obtained from the carotid artery of patients with atherosclerosis, and were isolated and placed in liquid nitrogen. Patients (male vs. female, 3:2; age, 57-68 years, mean age, 61.4 years) eligible for inclusion in the study were required to have a confirmation of pathological diagnosis of primary atherosclerosis, while those with metastatic tumors, congestive heart failure, diabetes mellitus, hematologic disease and/or communicable diseases were excluded. All patients were recruited from Nanfang Hospital, Guangzhuo, China between April 2017 and January 2020. The research study was approved by the Committee for Ethical Review of Research Involving Human Subjects, Nanfang Hospital, Southern Medical University, (Guangzhou, China; approval no. NFEC-2018-142). Identifying information of patients were not included in the study, so oral informed consent was obtained from the participants
Lentivirus construction and cell infection
RP11-531A24.3 cDNA was amplified by PCR and subsequently cloned into a CMV-MCS-PGK-Puro vector, and the correct sequence of the RP11-531A24.3 gene in this construct was validated through sequencing by Guangzhou RiboBio Co., Ltd. Empty lentiviral vector (LV-mock) and LV-lncRNA RP11-531A24.3 overexpression vector (LV-OE) was used to infect HA-VSMCs at a multiplicity of infection of 100 transducing units per cell in the presence of 8 mg/ml polybrene. A 1 µg/ml quantity of puromycin was added into the cell medium to select the stably transduced strain. Subsequent experiments were carried out following 6 months, at 10 generations after stable infection.
The siRNA was diluted as required. Before transfection, cells were incubated on a 6-well plate at a density of 1x105/well. On the next day, the cells were gently washed with PBS and 750 µl Opti-MEM was added to each well, and the cells were again incubated for 30 min. The working fluid containing 5 µl siRNA, 5 µl Lipofectamine 3000 and 240 µl Opti-MEM was mixed and reacted for 20 min. The working fluid was then added to the 6-well plate and cultured for 8 h, before replacing the transfection medium with fresh medium. The cells were divided into three groups: i) Cell group infected with si-HIF-1α or si-ANXA2; ii) Cell group infected with si-NC; and iii) Cell group used as a blank control. Subsequent experiments were carried out after 3 days. The siRNA sequences (Guangzhou RiboBio Co., Ltd.) are listed in Table I.
Table I.
siRNA sequences of ANXA2 and HIF-1α.
| siRNA | Sequence (5'-3') |
|---|---|
| si-ANXA2 | GTCTGTCAAAGCCTATACT |
| si-HIF-1α | CCAGCAACTTGAGGAAGTA |
ANXA2, annexin A2; HIF-1α, hypoxia inducible factor-1α; siRNA, small interfering RNA.
Cell cycle analysis
Cells were seeded at a density of 1x106 in 6-well plates. Following fixation with 70% ice-cold ethanol overnight at 4˚C, the samples were incubated with RNase A for 30 min at 37˚C and subsequently stained with propidium iodide for 10 min in room temperature. DNA in the labeled cells was identified using a flow cytometer (ModFit LT 3.1; BD Biosciences).
Cell apoptosis analysis
Cells were seeded at a density of 1x106 in 6-well plates. Samples were incubated in serum-free medium for 48 h, trypsinized and washed with cold PBS once, followed by Annexin V-APC staining for 10 min and 7-AAD staining for 5 min at room temperature. The apoptotic rate (both early-stage and late-stage apoptosis) was measured via flow cytometry (FlowJo 10; BD Biosciences).
Cell migration assay
Transwell chambers were used to assess cell migration. Briefly, cells were resuspended in serum-free medium at a density 2-4x104/ml. A total of 100-200 µl of the cell suspension (4x104/ml) was placed in the upper chamber and 600 µl of DMEM with 20% FBS was added to the lower chamber. The cells were incubated at 37˚C for 12 h. The cells from the upper surface of the filter were removed. Next, cells in the lower surface of the filter were fixed with methanol at room temperature for 30 min and stained with crystal violet at room temperature for 15 min. The stained cells were observed using a Nikon Eclipse Ts2R microscope in five randomly selected fields of each chamber (magnification, x200). Cell counts were determined using ImageJ v1.8.0 (National Institutes of Health).
Cell proliferation assay
Cell proliferation was evaluated using CCK-8. Cells were suspended in 96-well plates for 12 h for adhesion at a density of 2,000 cells/well (100 µl/well). At the beginning of this experiment, we had laid 4 identical 96-wells plates. After cell adhesion, CCK-8 solution (10 µl/well) was added to the medium for 2 h at 0, 12, 24 and 48 h time points, in four identical 96-wells plates, respectively. The absorbance was read at 450 nm using a microplate reader at each time point.
Total RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA from cultured cells and tissues was extracted using TRIzol reagent. The lncRNA RT was performed using an All-in-One First-Strand cDNA Synthesis kit in a 20-µl reaction mixture containing 2,000 ng total RNA. The reaction conditions for RT were as follows: 10 µl RNA was heated to 70˚C for 10 min for denaturation and mixed with 2 µl 10 mM dNTPs, 0.5 µl M-MLV reverse transcriptase, 0.5 µl M-MLV RNase inhibitor, 4 µl 5X RT buffer, 0.5 µl reverse transcription primer for RP11-531A24.3 or 0.5 µl reverse transcription primers for GAPDH (reference gene), and RNase-free dH2O. After mixing, the mixture was incubated at 42˚C for 60 min and at 85˚C for 5 min.
RT for mRNA used a PrimeScript II 1st Strand cDNA Synthesis kit in a 20-µl reaction mixture containing 2,000 ng total RNA. The reaction conditions were as follows: 4 µl 5X PrimeScript buffer, 1 µl PrimeScript RT Enzyme Mix, 1 µl Random 6 mers (100 µM), 1 µl Oligo dT Primer (50 µM), 13 µl of RNA and RNase-free dH2O; after uniform mixing, the mixture was incubated at 37˚C for 15 min and at 85˚C for 5 sec.
qPCR was performed using an SYBR Green PCR kit on an ABI 7500 Fast Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) after cDNA was diluted five times. The reaction system and the thermocycling conditions were as follows: 10 µl SuperMix, 0.5 µl forward primers, 0.5 µl reverse primers, 4 µl RNase-free dH2O and 5 µl cDNA were mixed, before enzyme activation at 50˚C for 2 min, followed by 45 cycles of 95˚C for 15 sec and 60˚C for 32 sec. Samples were then heated to 95˚C for 1 min, 60˚C for 30 sec and 40˚C for 30 sec. GAPDH served as the endogenous control for lncRNA and mRNA. Results were calculated using the 2-ΔΔCq method (24). The primers used in the analysis are listed in Table II.
Table II.
Primer sequences for RP11-531A24.3, ANXA2 and GAPDH
| Primer name | Sequence (5'-3') |
|---|---|
| RP11-531A24.3 | RT: ATTTAGTGGGCTGGTGGG |
| R: CAAATAACCACCCGAAAA | |
| F: ACCAAAGCAGATAGGCAC | |
| GAPDH | RT: TGGTGAAGACGCCAGTGGA |
| R: TGGTGAAGACGCCAGTGGA | |
| F: GCACCGTCAAGGCTGAGAAC | |
| ANXA2 | R: ATTTAGTGGGCTGGTGGG |
| F: CAAATAACCACCCGAAAA |
RT, reverse transcription; R, reverse; F, forward; ANXA2, annexin A2.
Western blot analysis
Cells with or without hypoxia culture were used for total protein extraction. Total protein was extracted using RIPA buffer. Protein concentrations were determined using a Nanodrop (NanoDrop 2000; Thermo Fisher Scientific, Inc.) and separated via 12% (ANXA2; 120 µg) or 10% (HIF-1α; 320 µg) SDS-PAGE before transfer to a PVDF membrane. The membrane was blocked with 5% bovine serum albumin for 1 h at room temperature and then incubated overnight at 4˚C with primary antibodies (ANXA2, 1:1,000; HIF-1α, 1:500; tubulin, 1:10,000). After incubation with the Goat anti-Rabbit IgG secondary antibody (1:10,000; cat. no. 31460; Thermo Fisher Scientific, Inc.) at room temperature for 30 min, immunoreactivity was detected by enhanced chemiluminescence using Enhanced Western Blotting Substrate. The protein expression levels were quantified with ImageJ software v1.8.0.
Fluorescence in situ hybridization (FISH)
A green fluorescence-labeled lncRNA probe recognizing RP11-531A24.3 was designed and synthesized according to the sequence analysis and design principles (BersinBio). The probe sequence is shown in Table III.
Table III.
Probe sequence for RP11-531A24.3 used for fluorescence in situ hybridization.
| Gene name | Probe sequence |
|---|---|
| ENSG0000564832 | CAGGCAGGTCTACATTGGCAATGGAAAATAAGCAATTATATGGGAAAATCAGTA GATGTTTCTCTTTAGTTTGTTAGTAGGCAACACTTTTAAACTGAATTACTCAATGTATTTTGAC TATGTAGATATGACACAGATATTTATTACAAGCCTGAAAAAACAGTTAAAAAATACTATTTCAG TATTTACGGTAAAGAATACACAGATGTAAATGATTCCAACAGTGAGCCAGTTTGACTAAAAGC GTTATTGCACTGCCTCAG |
Cells were fixed with 4% paraformaldehyde for 20 min at room temperature, permeabilized with transparent liquid (10% Triton X-100, 10X PBS, H2O) for 10 min at room temperature, refixed with 1% paraformaldehyde for 10 min and dehydrated using 70, 80, 95 and 100% ethanol for 5 min each. The cells were transferred rapidly to a hybridization system containing the probe and incubated overnight at 53˚C. DAPI (10 mg/ml) was added for 10 min to stain the nucleus at room temperature. The samples were imaged using a confocal laser microscope, and two fields of view were captured (magnification, x200).
RAP and liquid chromatography-mass spectrometry (LC-MS)
RAP was performed to separate lncRNA RP11-531A24.3 and its RBPs by designing biotin or streptomycin probes capable of binding to the target lncRNA fragments. A total of 10 antisense DNA probes (control group) and 10 sense DNA probes (experimental group, which were divided into odd probes, including number 1, 3, 5, 7 and 9, and even probes, including number 2, 4, 6, 8 and 10) with 5'-biotin labels were ordered and purchased from BersinBio. The probe sequences are shown in Table IV.
Table IV.
Probe sequences for RP11-531A24.3 used for RNA antisense purification.
| A, Odd probe | |
|---|---|
| Probe sequence (5'-3') | Probe location |
| ATTCACATAATGACTAAGCATACTCAAATGTGTGGTAACAAGGAGGC | 224-270 |
| CCTCAACAAACCAACCAAACAAATAAAATACCCCAAATAACCACCCGAAA | 917-966 |
| CCAGGCAGGTCTACATTGGCAATGGAAAATAAGCAATTATATGGGAAAA | 1,624-1,672 |
| TCCTCCACGCCAATCAACTTTTAGAGAATATACCATGCAAACTTTATTTA | 2,042-2,091 |
| AAAGTCAATAATGCTTCATGCGACTGTACCTAGAAATCTAATACCATGTATA | 2,600-2,651 |
| B, Even probe | |
| Probe sequence (5'-3') | Probe location |
| CTTGCTGCATTATATGTAGTCCTTGGTCATCACCCAGCTCCACTGCT | 627-673 |
| ATTTAATACATTTAGTGGGCTGGTGGGCAGCATAAGGCTTTATCTGAATACTATTTCA | 1,169-1,226 |
| TTCCATCCTTTCTTTTCCCTTCCCCACCAATGCTGGCCTT | 1,891-1,930 |
| ACTTTGTGAATCTCCACAGAGCTGTCCTGAGTTTAACAATAATCTTCTAA | 2,249-2,298 |
| TTTTCTATCTCGCTTATTCTACCAGACTGAAATGGAGAACAATGCCAGCAATTTTATA | 2,757-2,816 |
| C, Negative probe | |
| Probe sequence (5'-3') | Probe location |
| GAGCCACCACACCCAGCCCTTAAAATTATTAAGACTTCACATATACT | 154-200 |
| TTCATAACCAGTTTGGTTCTAGATTTACTGTAATTGGCTATCTGCCAGAATAAACT | 339-394 |
| GGGTGATGACCAAGGACTACATATAATGCAGCAAGCACGCT | 639-679 |
| TTATTTGGGGTATTTTATTTGTTTGGTTGGTTTGTTGAGGGGTTTT | 927-972 |
| AGATAAAGCCTTATGCTGCCCACCAGCCCACTAAATGTATTAAATACCTG | 1,182-1,231 |
| GTTTAAAAGTGTTGCCTACTAACAAACTAAAGAGAAACATCTACTGATTTTCCCAT | 1,577-1,632 |
| AAGGCCAGCATTGGTGGGGAAGGGAAAAGAAAGGATGGAA | 1,891-1,930 |
| AAACTCAGGACAGCTCTGTGGAGATTCACAAAGTAATTTCATG | 2,265-2,307 |
| TAAGTTATACATGGTATTAGATTTCTAGGTACAGTCGCATGAAGCATTATTGACTTT | 2,595-2,651 |
| ATATGAGTTAGCATACTCGTGTTTGTTCAGCTGTCCATCCTGCATCG | 2,962-3,008 |
Probe location of the odd and even groups refers to the binding site of those probes on RP11-531A24.3, but the probe location of the negative probe refers to the binding site of the negativeprobes on the antisense strand of RP11-531A24.3.
After performing cross-linking of cells, collection of the cell pellet, homogenization and DNA removal processes (25), the cell lysate was divided into the following three groups: RAP odd, RAP even and RAP NC, and the corresponding probes were added for hybridization (20 pmol probes; denaturation for 10 min at 65˚C, hybridization for 180 min at 45˚C). Magnetic beads were added to the three sample-probes for enrichment. A magnetic rack was used to collect the beads, and the supernatant was removed. Each group was divided into two parts: Protein detection and RNA detection. The RNA detection group was used to confirm whether the pulldown fragment of probes, including both odd and even groups, were from RP11-531A24.3. A total of 30 µl of each protein sample was collected for LC-MS detection.
The protein solution was subjected to the enzymatic digestion step. The peptide was dissolved and centrifuged with a sample solution (0.1% formic acid, 2% acetonitrile); the supernatant was transferred, and mass spectrometry was performed (Q Exactive; Thermo Fisher Scientific, Inc; positive; 320˚C; full scan mode; 350-1,800 m/z, 300 nl/min). The original mass spectrometry file was converted to an MGF file format, and the peptides were searched against the Uniprot database using MASCOT (http://www.matrixscience.com/).
Statistical analysis
Each experiment was performed in triplicate. Statistical analysis was performed using SPSS 22.0 program (IBM, Corp.). Statistical evaluation was carried out using Student's t-test for comparison between the two groups and ANOVA followed by the Bonferroni method was used for comparison of >2 groups. A paired t-test was used between atherosclerotic plaque tissues and normal intima tissues which were obtained from the same individuals. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of RP11-531A24.3 is reduced in advanced atherosclerotic lesions, and RP11-531A24.3 inhibits the proliferation and migration of HA-VSMCs
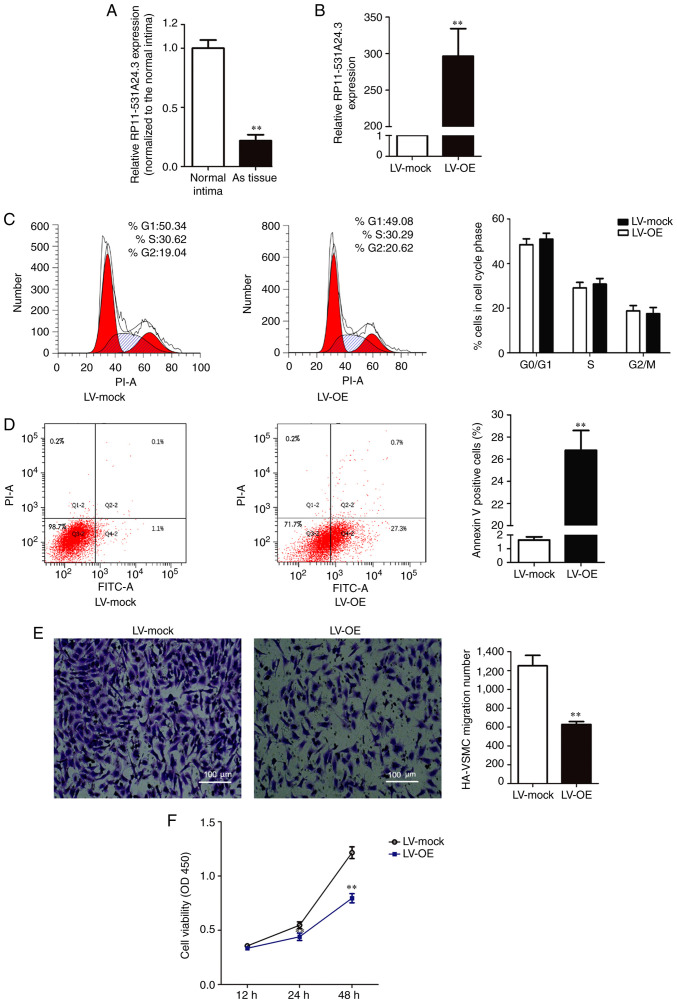
Microarray analysis indicated that RP11-531A24.3 (https://ensembl.org/, ENST00000564832.1) expression was reduced in the advanced atherosclerosis plaque, with a 222.5-fold reduction in expression compared with control tissues (n=3). RP11-531A24.3 was further confirmed as a non-coding or less coding transcript by the Coding Potential Calculator. To validate the varying expression levels of RP11-531A24.3, five paired atherosclerotic plaque tissues and normal intima tissues were examined via RT-qPCR (Fig. 1A). The results indicated that RP11-531A24.3 was downregulated in atherosclerotic plaque tissues compared with normal tissues (fold change =0.179).
Figure 1.
RP11-531A24.3 inhibits the proliferation and migration of HA-VSMCs. (A) RP11-531A24.3 expression was reduced in atherosclerotic plaque tissue in comparison with control tissues. (B) HA-VSMCs were transfected with LV-mock and LV-OE. (C) Flow cytometry analysis suggested that RP11-531A24.3 had no effect on cell cycle distribution. (D) Flow cytometry analysis suggested that RP11-531A24.3 increased cell apoptosis. (E) Transwell assay showed that RP11-531A24.3 reduced cell migration (scale bar, 100 µm). (F) Cell Counting Kit-8 assay showed that RP11-531A24.3 reduced cell viability. Data are presented as the mean ± SD (n=3) except data of (A) are presented as the mean ± SD (n=5). **P<0.01 vs. normal intima or LV-mock. HA-VSMCs, human aorta-vascular smooth muscle cells; LV-mock, empty lentiviral vector; LV-OE, lentiviral RP11-531A24.3 overexpression vector; As, atherosclerosis; PI, propidium iodide.
HA-VSMCs were infected with LV-mock or LV-OE (Fig. 1B). A series of gain-of-function studies were performed in HA-VSMCs to investigate the regulatory effect of RP11-531A24.4 on the phenotypes of VSMCs (Fig. 1C-F). The first study was conducted to investigate whether RP11-531A24.3 was involved in cell cycle distribution and apoptosis. The flow cytometry analysis showed no significant difference in the number of cells in the G0/G1 phase, S phase and G2/M phase between the LV-OE and LV-mock groups (Fig. 1C). The cell apoptosis test showed that the apoptotic rate was significantly increased in the LV-OE group compared with in LV-mock group (Fig. 1D). The Transwell assay indicated that overexpression of RP11-531A24.3 significantly inhibited the migration of HA-VSMCs compared with LV-mock (Fig. 1E). The CCK-8 assay showed that overexpression of RP11-531A24.3 reduced the viability of the cells in comparison with a control (Fig. 1F). Taken together, these results demonstrated that RP11-531A24.3 inhibited migration and increased apoptosis, while it had no effect on the cell cycle distribution of these cells.
RP11-531A24.3 is located in the cytoplasm of HA-VSMCs and is associated with the KEGG term ‘cytoskeletal regulation by Rho GTPase’
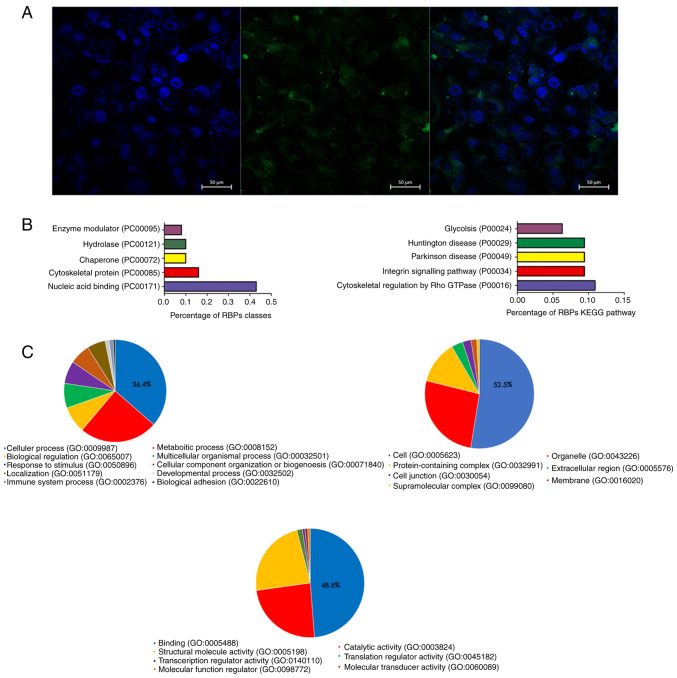
The cellular localization of RP11-531A24.3 in HA-VSMCs was assessed via FISH. The result indicated that RP11-531A24.3 was localized in the cytoplasm of HA-VSMCs (Fig. 2A).
Figure 2.
RP11-531A24.4 is located in the cytoplasm of HA-VSMCs and its expression is associated with the KEGG term ‘cytoskeletal regulation by Rho GTPase’. (A) Representative images of RP11-531A24.3 cellular localization obtained by fluorescence in situ hybridization. DAPI (blue fluorescence) stained the nucleus and RP11-531A24.3 is indicated by green fluorescence localized to the cytoplasm. (B) Left: the protein classification of associated RBPs. Right: Pathways with the highest proportion of associated RBPs in the KEGG pathway enrichment analysis. (C) GO analysis of RBPs was performed for the biological process, cellular component and molecular function categories (left side, right side, the center). Pie charts represent the percentage of the analyzed RBPs associated with each term. HA-VSMCs, human aorta-vascular smooth muscle cells; KEGG, Kyoto Encyclopedia of Genes and Genomes; RBP, RNA-binding protein; GO, Gene Ontology.
The RAP-MS experimental results showed that there were 141 proteins that bound with RP11-531A24.3 directly. KEGG pathway enrichment (Fig. 2B) and GO analysis (Fig. 2C) were conducted to investigate the biological function of those RBPs. The most commonly associated terms were ‘nucleic acid binding’, ‘cytoskeletal protein’, ‘hydrolase’ and ‘chaperone’ (Fig. 2B). The top three molecular function terms from the GO analysis of the RBPs were ‘binding (GO:0005488)’, ‘catalytic activity (GO:0003824)’ and ‘structural molecule activity (GO:0005198)’ (Fig. 2C). ‘Cytoskeletal regulation by Rho GTPase (P00016)’ ranked the highest in the enrichment analysis of the KEGG pathway (Fig. 2B). Translocation of VSMCs is driven by a dynamic reorganization of the actin cytoskeleton (26). The regulatory mechanism of actin recombination is unclear; however, it is thought that complex interactions between actin cytoskeleton-related proteins and GTPases may play a role in this mechanism (26,27).
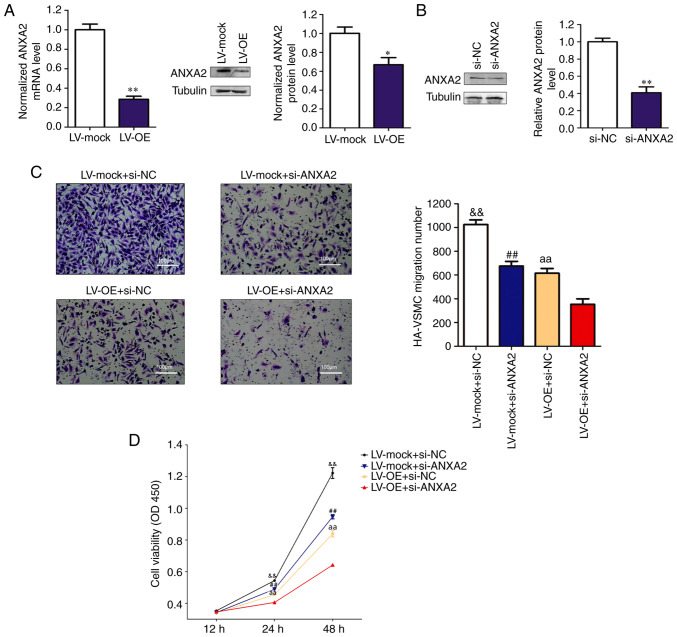
RP11-531A24.3 suppresses the migration and proliferation of HA-VSMCs via inhibition of ANXA2 expression
Among the RBPs, ANXA2, whose enhanced expression has been reported to potentiate the migration, invasion and proliferation of various cancers (28), was identified. ANXA2 is a member of the calcium-dependent phospholipid-binding protein family and is related to the smooth muscle contraction pathway (29,30). A previous study using Affinity Capture-MS showed that ANXA2 binds to myosin heavy chain 9 protein, which is one of the downstream proteins in the Rho GTPase regulatory pathway (31). Overexpression of RP11-531A24.3 suppressed ANXA2 expression at both mRNA and protein levels compared with controls (Fig. 3A). The reduced migration and viability mediated by RP11-531A24.3 overexpression were more significantly suppressed after ANXA2 depletion in RP11-531A24.3-overexpressing cells compared with in controls (Fig. 3B-D). Collectively, these data suggested that RP11-531A24.3-mediated depletion of the expression and function of ANXA2 inhibited the migration and viability of HA-VSMCs.
Figure 3.
RP11-531A24.3 suppresses the migration and proliferation of HA-VSMCs via inhibition of ANXA2 expression. (A) Overexpression of RP11-531A24.3 suppresses ANXA2 expression at both the mRNA and protein levels. (B) ANXA2 knockdown by siRNA in HA-VSMCs was examined via western blot analysis. (C) Transwell and (D) Cell Counting Kit-8 assays were used to evaluated the migration and viability of HA-VSMCs co-transfected with LV-mock, LV-OE, si-NC or si-ANXA2 (scale bar, 100 µm). Data are presented as the mean ± SD (n=3). *P<0.05; **P<0.01 vs. LV-mock; aaP<0.01 vs. LV-OE+si-NC; ##P<0.01 vs. LV-mock + si-ANXA2; &&P<0.01 vs. LV-mock + si-NC. HA-VSMCs, human aorta-vascular smooth muscle cells; ANXA2, annexin A2; LV-mock, empty lentiviral vector; LV-OE, lentiviral RP11-531A24.3 overexpression vector; si, small interfering; NC, negative control.
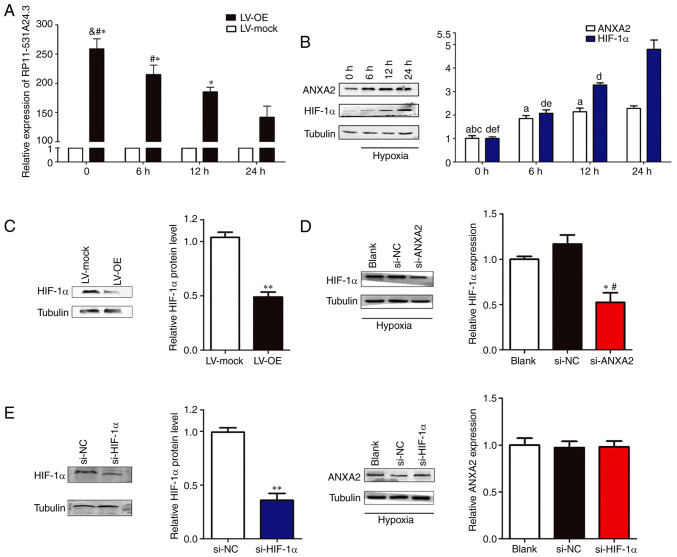
Hypoxia regulates the expression of RP11-531A24.3 and ANXA2 in HA-VSMCs
HA-VSMCs transfected with LV-OE or LV-mock and normal HA-VSMCs were cultured in normal conditions (20% O2) or hypoxic conditions (1% O2) for 6-24 h. Next, the expression levels of RP11-531A24.3, ANXA2 and HIF-1α were analyzed. The results indicated that the expression of RP11-531A24.3 was reduced (Fig. 4A), while that of ANXA2 and HIF-1α was augmented, by hypoxia compared with controls (Fig. 4B). Of note, overexpression of RP11-531A24.3 suppressed HIF-1α expression at the protein level (Fig. 4C). Under hypoxic conditions, transfection with siRNA targeting HIF-1α exerted no significant effects on ANXA2 expression, while an siRNA targeting ANXA2 decreased the levels of HIF-1α expression in HA-VSMCs (Fig. 4D and E). These results indicated that hypoxia may regulate the expression of RP11-531A24.3 and ANXA2, and facilitate the expression of HIF-1α via ANXA2.
Figure 4.
Hypoxia regulates the expression of RP11-531A24.3, ANXA2 and HIF-1α in HA-VSMCs. Effect of hypoxia on the expression of (A) RP11-531A24.3 *P<0.05 vs. 24 h; #P<0.05 vs. 12 h; &P<0.05 vs. 6 h and (B) ANXA2, aP<0.05 vs. 24 h; bP<0.05 vs. 12 h; cP<0.05 vs. 6 h; and HIF-1α, dP<0.05 vs. 24 h; eP<0.05 vs. 12 h; fP<0.05 vs. 6 h. (C) HIF-1α expression at protein levels under hypoxic conditions was detected in HA-VSMCs transfected with LV-OE or LV-mock. **P<0.01 vs. LV-mock. (D) HIF-1α expression at the protein level was detected in HA-VSMCs transfected with si-NC, si-ANXA2 and blank control. *P<0.05 vs. si-NC; #P<0.05 vs. Blank. (E) HIF-1α and ANXA2 expression levels were detected in HA-VSMCs transfected with si-NC, si-HIF-1α and blank control. **P<0.01 vs. si-NC. Data are presented as the mean ± SD (n=3). HA-VSMCs, human aorta-vascular smooth muscle cells; ANXA2, annexin A2; HIF-1α, hypoxia inducible factor-1α; LV-mock, empty lentiviral vector; LV-OE, lentiviral RP11-531A24.3 overexpression vector; si, small interfering; NC, negative control.
Discussion
The major clinical consequences of atherosclerosis are not due to the diminishing blood supply in the gradually narrowing lumen caused by stable plaques, but rather due to a sudden blockage of blood flow caused by acute rupture or decomposition of an unstable plaque in a thrombotic event (5,16). The development of new therapies to treat atherosclerosis and minimize its clinical consequences will be contingent on a rigorous understanding of the biology of VSMCs, which contribute to the pathogenesis of advanced lesions. In advanced lesions, plaque disruption is inversely proportional to the number of VSMCs, which is closely related to the proliferation, migration, and death of VSMCs (4). In humans, advanced atherosclerotic lesions demonstrate little VSMC proliferation (4,5,16). The proliferation of VSMCs in the development of atherosclerosis appears to be predominantly restorative, rather than as a major driver of plaque formation (5). Human VSMCs can migrate in response to varying stimuli in vitro; however, it remains unclear how VSMC migration affects atherosclerosis and whether migration occurs independent of proliferation (4,5). In present study, RP11-531A24.3 was identified through screening of differentially expressed genes in an lncRNA microarray in three advanced atherosclerosis samples and in three normal intima tissue. Next, a series of gain-of-function assays were performed to demonstrate that RP11-531A24.3 was associated with migration and proliferation phenotypes in HA-VSMCs. KEGG pathway analysis indicated that the RBPs were enriched in the pathway termed ‘cytoskeletal regulation by Rho GTPase’, which has been reported to be closely associated with cell migration and invasion in tumor cell metastasis (32).
ANXA2 is one of the RBPs of RP11-531A24.3, and is a member of the calcium-dependent phospholipid-binding protein family. ANXA2 plays a key role in tumorigenesis and cancer progression, and is involved in the positive regulation of low-density lipoprotein receptor activity, lipoprotein receptor binding and lipoprotein clearance (28-30,33,34). Given the biological function of ANXA2 in cancers and atherosclerosis (28,29,33,34) and based on the association between RP11-531A24.3 and ANXA2, it was investigated and demonstrated that RP11-531A24.3 inhibited the migration and proliferation of HA-VSMCs through binding to ANXA2 directly in the cytoplasm to reduce its expression at both mRNA and protein levels. Collectively, the results of the present study may advance understanding of the biological behavior of lncRNA RP11-531A24.3 in HA-VSMCs, and may serve as a preliminarily investigation into the mechanism by which RP11-531A24.3 regulates the migration and proliferation of VSMCs via ANXA2.
Non-coding RNAs, including microRNAs, circular RNAs and lncRNAs, are considered to serve central roles in the regulation of phenotypic transformation of SMCs, involving changes in migration, proliferation, apoptosis, and ECM-generating capacity (16,35,36). The subcellular localization of lncRNA is likely to define its network of interactions with other macromolecules and their functions (15). In the present study, FISH indicated the cytoplasmic localization of RP11-531A24.3, while RAP and LC-MS indicated its RBPs were enriched in the pathway terms closely related to migration. These results were consistent with phenotypic changes caused by RP11-531A24.3.
ANXA2, a Ca2+-dependent phospholipid-binding protein that directly binds to RP11-531A24.3, has a unique N-terminus tail domain that contains a nuclear export signal and multiple phosphorylation sites (28,29). It forms a heterotetramer and drives the conversion of plasminogen to plasmin at the cell surface (33,37); this further activates metalloproteinases and degrades EMC components, which is an essential process for cell metastatic progression (28,37). ANXA2 is upregulated in various cancers and is related to cell angiogenesis, proliferation, invasion, and adhesion (28,30). Upregulation of ANXA2 is reported to be related to a more proangiogenic phenotype in macrophages and more migratory phenotype in VSMCs (34,38). In the present study, a direct interaction between ANXA2 and RP11-531A24.3 that regulates the proliferation and migration of HA-VSMCs was indicated. However, the binding site and the binding mechanism need to be further investigated.
The anoxemia hypothesis proposes that an unbalanced supply and demand for oxygen in the arterial wall is one of the key factors in the development of atherosclerosis (9,10). The presence of hypoxic areas in developing atherosclerotic plaques has been widely reported to alter the function, metabolism, and responses of various cell types, and to determine whether the plaque will evolve into an unstable phenotype (9,11). In SMCs, HIF-1α plays an important role in cell proliferation and migration, two important features of foam cell formation, and also mediates multiple mechanisms, such as macrophage outflow pathways (10). In esophageal squamous cell carcinoma, phosphorylated ANXA2 inhibited ubiquitin-dependent proteasomal degradation of MYC proto-oncogene protein, which potentiated HIF-1α transcription and promoted migration, invasion, and metastasis (30). ANXA2 expression was delineated in RAW264.7 macrophages under hypoxic condition (34). The present study indicated that the expression levels of ANXA2 and HIF-1α were markedly augmented in hypoxic conditions and that knockdown of ANXA2 decreased HIF-1α expression, but the mechanism related to these aspects remain to be elucidated. lncRNAs are also involved in hypoxia-induced pulmonary hypertension-related vascular remodeling by acting as a sponge or through the regulation of genes involved in cell proliferation, migration and apoptosis in pulmonary VSMCs (39). The results of the present study demonstrated that lncRNA RP11-531A24.3 was downregulated in HA-VSMCs in hypoxia. Further studies on how hypoxia manipulates RP11-531A24.3, as well as the interaction between RP11-531A24.3 and ANXA2, are required in the future.
In conclusion, the present study suggested that lncRNA RP11-531A24.4 regulated the phenotypes of migration and proliferation in HA-VSMCs through ANXA2. The effect of hypoxia on the expression levels of RP11-531A24.3, ANXA2 and HIF-1α was assessed, however, HIF-1α expression was not identified by RAP-MS. It may be hypothesized that HIF-1α was not easy to detect due to its low expression abundance in the cytoplasm under a constant oxygen environment. Further research is required into how RP11-531A24.3 plays a role in phenotypic switching of VSMCs during atherosclerosis and the mechanism via which hypoxia regulates these molecules. This research may facilitate the development of new therapeutic strategies for the treatment of cardiovascular diseases.
Acknowledgements
Not applicable.
Funding Statement
Funding: This study was supported by grants from the National Natural Science Foundation of China (grant no. 81772244) and the Natural Science Fund of Guangdong (grant no. 2020B1515020013).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YW carried out the experiments and wrote the manuscript. FC and YL performed the statistical analysis and created the figures. QW and YH designed the study and revised the manuscript. YW and YL confirmed the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Committee for Ethical Review of Research Involving Human Subjects, Nanfang Hospital, Southern Medical University, Guangzhou, China (approval no. NFEC-2018-142). Oral informed consent was obtained from the participants or their relatives.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Roth GA, Johnson C, Abajobir A, Abd-Allah F, Abera SF, Abyu G, Ahmed M, Aksut B, Alam T, Alam K, et al. Global, Regional, and National Burden of Cardiovascular Diseases for 10 Causes, 1990 to 2015. J Am Coll Cardiol. 2017;70:1–25. doi: 10.1016/j.jacc.2017.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song P, Zha M, Yang X, Xu Y, Wang H, Fang Z, Yang X, Xia W, Zeng C. Socioeconomic and geographic variations in the prevalence, awareness, treatment and control of dyslipidemia in middle-aged and older Chinese. Atherosclerosis. 2019;282:57–66. doi: 10.1016/j.atherosclerosis.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Lusis AJ. Atherosclerosis. Nature. 2000;407:233–241. doi: 10.1038/35025203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett MR, Sinha S, Owens GK. Vascular Smooth Muscle Cells in Atherosclerosis. Circ Res. 2016;118:692–702. doi: 10.1161/CIRCRESAHA.115.306361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basatemur GL, Jørgensen HF, Clarke MCH, Bennett MR, Mallat Z. Vascular smooth muscle cells in atherosclerosis. Nat Rev Cardiol. 2019;16:727–744. doi: 10.1038/s41569-019-0227-9. [DOI] [PubMed] [Google Scholar]
- 6.Williams KJ, Tabas I. The response-to-retention hypothesis of early atherogenesis. Arterioscler Thromb Vasc Biol. 1995;15:551–561. doi: 10.1161/01.atv.15.5.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tabas I, Williams KJ, Borén J. Subendothelial lipoprotein retention as the initiating process in atherosclerosis: Update and therapeutic implications. Circulation. 2007;116:1832–1844. doi: 10.1161/CIRCULATIONAHA.106.676890. [DOI] [PubMed] [Google Scholar]
- 8.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 9.Ferns GAA, Heikal L. Hypoxia in Atherogenesis. Angiology. 2017;68:472–493. doi: 10.1177/0003319716662423. [DOI] [PubMed] [Google Scholar]
- 10.Jain T, Nikolopoulou EA, Xu Q, Qu A. Hypoxia inducible factor as a therapeutic target for atherosclerosis. Pharmacol Ther. 2018;183:22–33. doi: 10.1016/j.pharmthera.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Luque A, Turu M, Juan-Babot O, Cardona P, Font A, Carvajal A, Slevin M, Iborra E, Rubio F, Badimon L, et al. Overexpression of hypoxia/inflammatory markers in atherosclerotic carotid plaques. Front Biosci. 2008;13:6483–6490. doi: 10.2741/3168. [DOI] [PubMed] [Google Scholar]
- 12.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. ENCODE Project Consortium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mishra K, Kanduri C. Understanding Long Noncoding RNA and Chromatin Interactions: What We Know So Far. Noncoding RNA. 2019;5(5) doi: 10.3390/ncrna5040054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDonel P, Guttman M. Approaches for Understanding the Mechanisms of Long Noncoding RNA Regulation of Gene Expression. Cold Spring Harb Perspect Biol. 2019;11(11) doi: 10.1101/cshperspect.a032151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sauvageau M. Diverging RNPs: Toward Understanding lncRNA-Protein Interactions and Functions. Adv Exp Med Biol. 2019;1203:285–312. doi: 10.1007/978-3-030-31434-7_10. [DOI] [PubMed] [Google Scholar]
- 16.Leeper NJ, Maegdefessel L. Non-coding RNAs: Key regulators of smooth muscle cell fate in vascular disease. Cardiovasc Res. 2018;114:611–621. doi: 10.1093/cvr/cvx249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar S, Williams D, Sur S, Wang JY, Jo H. Role of flow-sensitive microRNAs and long noncoding RNAs in vascular dysfunction and atherosclerosis. Vascul Pharmacol. 2019;114:76–92. doi: 10.1016/j.vph.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holdt LM, Kohlmaier A, Teupser D. Long Noncoding RNAs of the Arterial Wall as Therapeutic Agents and Targets in Atherosclerosis. Thromb Haemost. 2019;119:1222–1236. doi: 10.1055/s-0039-1692680. [DOI] [PubMed] [Google Scholar]
- 19.Xu S, Kamato D, Little PJ, Nakagawa S, Pelisek J, Jin ZG. Targeting epigenetics and non-coding RNAs in atherosclerosis: From mechanisms to therapeutics. Pharmacol Ther. 2019;196:15–43. doi: 10.1016/j.pharmthera.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu YW, Guo FX, Xu YJ, Li P, Lu ZF, McVey DG, Zheng L, Wang Q, Ye JH, Kang CM, et al. Long noncoding RNA NEXN-AS1 mitigates atherosclerosis by regulating the actin-binding protein NEXN. J Clin Invest. 2019;129:1115–1128. doi: 10.1172/JCI98230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai HL, Lu ZF, Zhao JJ, Ma X, Li XH, Xu H, Wu SG, Kang CM, Lu JB, Xu YJ, et al. Microarray profiling analysis and validation of novel long noncoding RNAs and mRNAs as potential biomarkers and their functions in atherosclerosis. Physiol Genomics. 2019;51:644–656. doi: 10.1152/physiolgenomics.00077.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49D:D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gene Ontology C. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021;49D:D325–D334. doi: 10.1093/nar/gkaa1113. Gene Ontology Consortium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Nan A, Chen L, Zhang N, Liu Z, Yang T, Wang Z, Yang C, Jiang Y. A novel regulatory network among lncRpa, circRar1, miR-671 and apoptotic genes promotes lead-induced neuronal cell apoptosis. Arch Toxicol. 2017;91:1671–1684. doi: 10.1007/s00204-016-1837-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukata Y, Amano M, Kaibuchi K. Rho-Rho-kinase pathway in smooth muscle contraction and cytoskeletal reorganization of non-muscle cells. Trends Pharmacol Sci. 2001;22:32–39. doi: 10.1016/s0165-6147(00)01596-0. [DOI] [PubMed] [Google Scholar]
- 27.Webb RC. Smooth muscle contraction and relaxation. Adv Physiol Educ. 2003;27:201–206. doi: 10.1152/advan.00025.2003. [DOI] [PubMed] [Google Scholar]
- 28.Lokman NA, Ween MP, Oehler MK, Ricciardelli C. The role of annexin A2 in tumorigenesis and cancer progression. Cancer Microenviron. 2011;4:199–208. doi: 10.1007/s12307-011-0064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayer G, Poirier S, Seidah NG. Annexin A2 is a C-terminal PCSK9-binding protein that regulates endogenous low density lipoprotein receptor levels. J Biol Chem. 2008;283:31791–31801. doi: 10.1074/jbc.M805971200. [DOI] [PubMed] [Google Scholar]
- 30.Ma S, Lu CC, Yang LY, Wang JJ, Wang BS, Cai HQ, Hao JJ, Xu X, Cai Y, Zhang Y, et al. ANXA2 promotes esophageal cancer progression by activating MYC-HIF1A-VEGF axis. J Exp Clin Cancer Res. 2018;37(183) doi: 10.1186/s13046-018-0851-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hussain S, Saxena S, Shrivastava S, Mohanty AK, Kumar S, Singh RJ, Kumar A, Wani SA, Gandham RK, Kumar N, et al. Gene expression profiling of spontaneously occurring canine mammary tumours: Insight into gene networks and pathways linked to cancer pathogenesis. PLoS One. 2018;13(e0208656) doi: 10.1371/journal.pone.0208656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang Y, He Y, Zhang P, Wang J, Fan C, Yang L, Xiong F, Zhang S, Gong Z, Nie S, et al. lncRNAs regulate the cytoskeleton and related Rho/ROCK signaling in cancer metastasis. Mol Cancer. 2018;17(77) doi: 10.1186/s12943-018-0825-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saiki Y, Horii A. Multiple functions of S100A10, an important cancer promoter. Pathol Int. 2019;69:629–636. doi: 10.1111/pin.12861. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z, Wei Q, Han L, Cao K, Lan T, Xu Z, Wang Y, Gao Y, Xue J, Shan F, et al. Tenascin-c renders a proangiogenic phenotype in macrophage via annexin II. J Cell Mol Med. 2018;22:429–438. doi: 10.1111/jcmm.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding P, Ding Y, Tian Y, Lei X. Circular RNA circ_0010283 regulates the viability and migration of oxidized low density lipoprotein induced vascular smooth muscle cells via an miR 370 3p/HMGB1 axis in atherosclerosis. Int J Mol Med. 2020;46:1399–1408. doi: 10.3892/ijmm.2020.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan Z, Wang H, Liang J, Li Y, Li X. MicroRNA-503-5p improves carotid artery stenosis by inhibiting the proliferation of vascular smooth muscle cells. Exp Ther Med. 2020;20(85) doi: 10.3892/etm.2020.9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deryugina EI, Quigley JP. Cell surface remodeling by plasmin: A new function for an old enzyme. J Biomed Biotechnol. 2012;2012(564259) doi: 10.1155/2012/564259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li L, Li X, The E, Wang LJ, Yuan TY, Wang SY, Feng J, Wang J, Liu Y, Wu YH, et al. Low expression of lncRNA-GAS5 is implicated in human primary varicose great saphenous veins. PLoS One. 2015;10(e0120550) doi: 10.1371/journal.pone.0120550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu B, Gong Y, Yan G, Wang D, Qiao Y, Wang Q, Liu B, Hou J, Li R, Tang C. Down-regulation of lncRNA MEG3 promotes hypoxia-induced human pulmonary artery smooth muscle cell proliferation and migration via repressing PTEN by sponging miR-21. Biochem Biophys Res Commun. 2018;495:2125–2132. doi: 10.1016/j.bbrc.2017.11.185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.