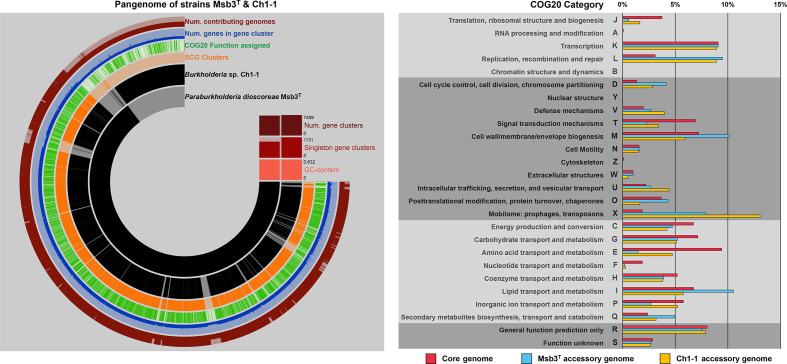
Fig. 2.
Pangenome analysis of strains Msb3T and Ch1-1. The left panel displays the pangenome of both strains created through ANVIO’s pangenomics suite [34]. CDSs from strain Msb3T and strain Ch1-1, respectively, are shown on ring 1 and 2 (starting inside). Presence of a CDS in either genome is indicated in black and absence in light grey. CDSs are forced into synteny with the genome sequence of strain Msb3T. Ring 3 displays whether two orthologs fall into self-consistency-grouping (SCG) clusters, ring 4, whether a COG20 function could be assigned to the CDS in the cluster. Ring 5 displays the number of genes that fall into a respective gene cluster, ranging from a minimum of one to a maximum of five, in form of a bar chart. The outermost ring, ring 6, indicates, how many genomes contribute to a gene cluster. If two genomes (full bar) contribute to a cluster, it was considered part of the core genome of the two strains. If, however, only one genome contributes (half bar), the respective CDS is considered part of that genome’s accessory genome (AG). The panel on the right shows the relative COG category classification patterns of the core genome of both strains in pink, the AG of strain Msb3T in light blue as well as the AG of strain Ch1-1 in yellow. The functional annotation into COG categories was conducted through the software package ANVIO, which implements the latest update of the COG database, COG20 [35]. Depicted is the percentage of all classifiable CDSs that fall into a given COG category.