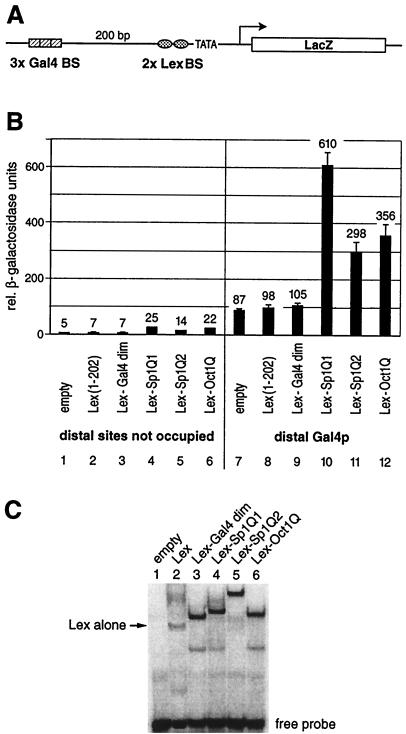
FIG. 2.
Glutamine-rich activation domains stimulate transcription from a chromosomally embedded reporter gene in yeast. (A) Schematic drawing of the integrated yeast reporter. The promoter architecture resembles that of a typical higher eukaryote. It contains distant (three Gal4 binding sites) as well as proximal (two Lex binding sites) regulatory elements upstream of the GAL1 TATA box. The reporter was integrated at the chromosomal URA3 locus of S. cerevisiae. (B) The reporter strain was transformed with yeast plasmids that express Lex fusion proteins to the Gal4 dimerization domain (amino acids 58 to 97) as a negative control and the same glutamine-rich activation domains of Sp1 (Q1 and Q2) and Oct1Q as tested in HeLa cells (Fig. 1). Their ability to stimulate transcription when bound to the proximal binding sites either alone or in combination with a distal Gal4p activator was monitored. The factors containing a glutamine-rich activation domain displayed a low intrinsic activation potential when tested on their own (lanes 4 to 6), which was above the background as determined by the Lex (lane 2) or Lex-Gal4 dimerization domain alone (lane 3). The influence on reporter gene activation of the distal Gal4p (lane 7) and the proximal glutamine-rich activation domains (lanes 4 to 6) was more than additive when tested in combination, i.e., they resulted in synergistic gene activation (lanes 10 to 12). As in the mammalian cells shown in Fig. 1, Sp1Q1 (lanes 4 and 10) was the strongest activation domain, followed by Oct1Q (lanes 6 and 12) and Sp1Q2 (lanes 5 and 11). (C) EMSAs were performed with total yeast protein extracts derived from reporter strains that expressed various Lex fusion proteins. A 32P-labeled oligonucleotide duplex with two Lex binding sites was used as a probe. Extracts from reporter strains without a Lex fusion protein did not reveal any protein interaction with the oligonucleotide (lane 1). Lex alone (lane 2) produced a weaker bandshift than the other fusion proteins. Fusion of the Gal4 dimerization domain to the Lex protein (lane 3) increased the stability of the protein to interact with the oligonucleotide and yielded a bandshift comparable to those of Lex fused to Sp1Q1 (lane 4), Sp1Q2 (lane 5), and Oct1Q (lane 6).