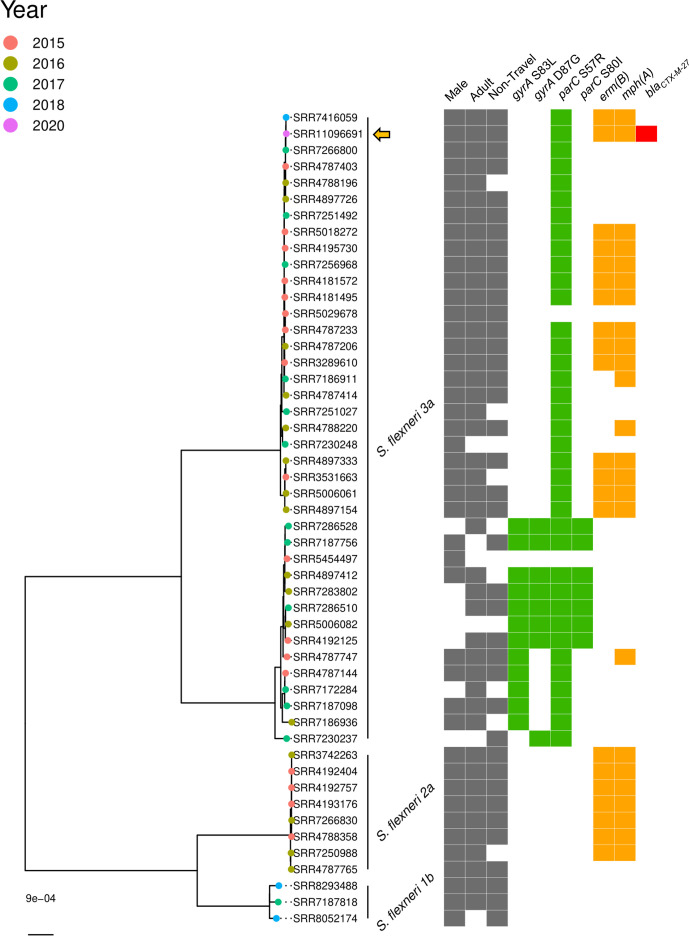
Fig. 3.
Maximum-likelihood phylogeny of S. flexneri 3a isolate 888 048 (SRR11096691, indicated by a filled orange arrow) sequenced with Nanopore technologies in this study, compared to other S. flexneri isolates in Public Health England’s collections. To place the isolate in context, 49 other S. flexneri isolates that are representatives from relevant SNP (single nucleotide polymorphism) clusters were included in the comparison. MSM clades associated with S. flexneri 1b, S. flexneri 2a and S. flexneri 3a are labelled. Isolates are labelled by SRR number and tip nodes are coloured by year of receipt. Adjacent grey blocks in each of the first three columns indicate the sample was isolated from a male, an adult, and that the illness was non-travel related (left-right). Grey blocks in these three lanes for an isolate implies the infection may be associated with MSM transmission. Presence of antimicrobial resistance genes concerning fluoroquinolones (mutations in gyrA and parC, green) and macrolides (erm(B) and mph(A), orange) are taken from Bardsley et al. [45] and presence of blaCTX-M-27 was determined using GeneFinder. Gene presence is indicated by a coloured tile in the relevant column. ML trees are midpoint rooted and scale bar indicates SNPs.