Fig. 4. Structure of iboxamycin (IBX) bound to the Erm-methylated 70S ribosome.
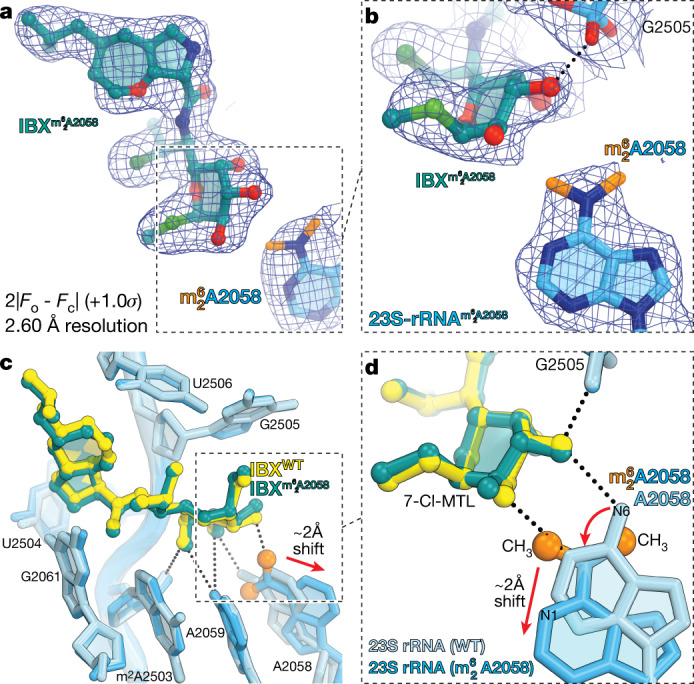
a, b, Electron density map (blue mesh), contoured at 1.0σ, of IBX (teal) in complex with the Erm-modified T. thermophilus 70S ribosome-containing N6-dimethylated A2058 residue in the 23S rRNA (a), highlighting the interaction with the methyl groups of (orange) (b). c, d, Superposition of IBX (yellow) in complex with the WT 70S ribosome containing an unmodified residue A2058 (light blue), and the structure of IBX (teal) in complex with the Erm-modified 70S ribosome containing an residue (medium blue) (c), highlighting hydrogen bonding (dashed lines) (d). Note that the position of IBX is almost identical in the two structures, whereas IBX binding to the Erm-modified ribosome causes substantial movement of from its canonical position (red arrow). WT, wild type.
