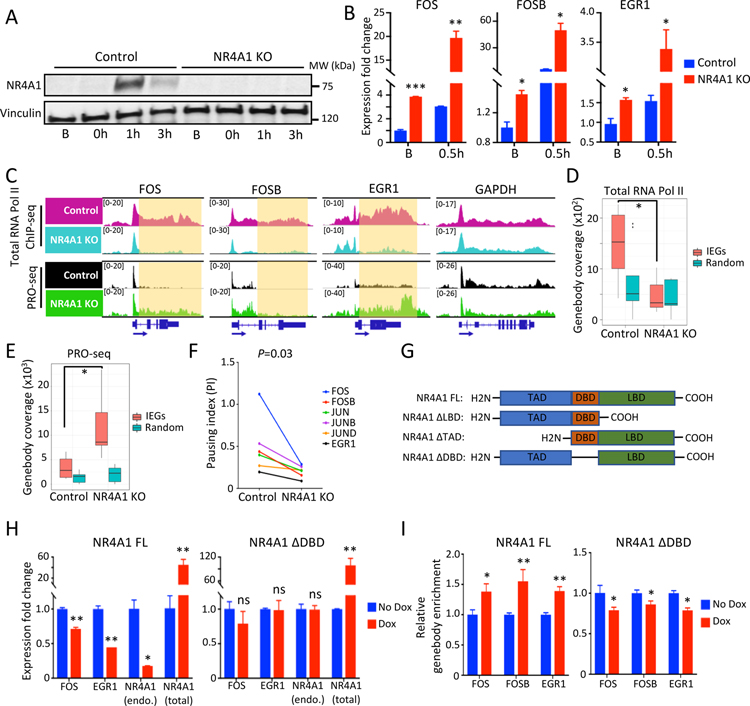
Figure 3. NR4A1 restrains IEG transcriptional elongation under unstressed conditions.
(A) Western blot shows absence of serum stress-induced NR4A1 in M231 with CRISPR-mediated NR4A1 deletion. Vinculin, loading control. (B) Increased IEG expression (FOS, FOSB, EGR1) in NR4A1-null M231, versus parental controls, at baseline (B) and upon serum replenishment of starved cells (0.5h). Error bar, SD. (C) IGV tracks show increased nuclear run-on signal (PRO-seq) across genebody of IEGs (FOS, FOSB, EGR1) in NR4A1-null M231 at baseline compared with parental M231. GAPDH, control locus. (D) Quantitation of reduced RNA Pol II ChIP-seq signal at genebodies of 6 IEGs (FOS, FOSB, JUN, JUNB, JUND and EGR1) in NR4A1-null cells, compared with parental cells. No difference in RNA Pol II genebody coverage is observed following NR4A1 deletion at 6 randomly selected genomic loci. (E) Increased PRO-seq coverage at 6 IEG genebodies in NR4A1-null versus parental cells. No increase is evident at 6 randomly selected genebodies. (F) Reduction in calculated RNA Pol II Pausing Index (PI) at six IEGs in NR4A1-null M231 versus parental controls. P=0.03, by Wilcoxon test. (G) Design of NR4A1 deletion constructs without transactivation domain (TAD), DNA binding domain (DBD), or ligand binding domain (LBD), compared with the full length (FL) construct. (H) Suppression of deregulated IEG mRNA expression (FOS, EGR1) in NR4A1-null M231, following inducible (Dox) re-expression of NR4A1-FL, but not NR4A1-ΔDBD, despite comparable expression. Error bar, SD. (I) ChIP-qPCR quantitation showing enrichment of NR4A1 binding at IEG genebodies (FOS, FOSB, EGR1) in NR4A1-null M231, following inducible (Dox) re-expression of NR4A1-FL, but not NR4A1-ΔDBD. In panels B, D, E, H and I, *P<0.05, **P<0.01, ***P<0.001, ns, not significant, by two-tailed Student’s t-Test.