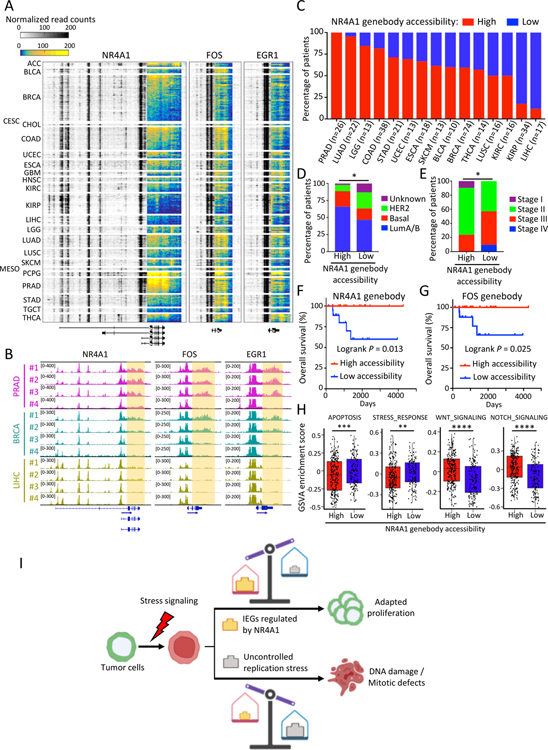
Figure 7. Prevalence of NR4A1 and IEG genebody chromatin accessibility in primary human cancers.
(A) Heatmap shows ATAC-seq signal at three IEG loci across TCGA tumor types, highlighting the prevalence of accessible chromatin at genebody and 3’-UTR regions. Color key from either blue to yellow (genebody) or white to black (other loci) indicates normalized read density from low to high. Original ATAC-seq data derived from (Corces et al., 2018). TCGA tumor study abbreviations are referenced from https://gdc.cancer.gov/resources-tcga-users/tcga-code-tables/tcga-study-abbreviations. (B) Representative IGV tracks show normalized ATAC-seq signal at IEG genebody (highlighted) in prostate (PRAD), breast (BRCA) and liver (LIHC) cancers (Corces et al., 2018). (C) Percentage of patients shown in (A) with primary tumors showing high (red bars) or low (blue bars) ATAC-seq signal at the NR4A1 genebody across TCGA cancer types. Only cancer types with more than 10 ATAC-seq samples are shown. Patients are classified into high and low groups according to the NR4A1 genebody/promoter ATAC-seq signal ratio (see Methods). A higher prevalence of favorable breast cancer histology (PAM50 molecular subtypes) (D) and lower pathologic tumor stages (AJCC) (E) is evident in patients with high NR4A1 genebody ATAC-seq signal. *P<0.05, **P<0.01 by Fisher exact test. (F-G) Kaplan-Meier plot showing improved overall survival in breast cancer patients with high accessibility (genebody ATAC-seq signal) at NR4A1 (panel F) and FOS (panel G). Statistical significance assessed by logrank test. (H) Box plots showing GSVA analysis of different pathways in all analyzed TCGA primary tumors (Corces et al., 2018) with high (n=237) or low (n=140) NR4A1 genebody accessibility. **P<0.01; ***P<0.001, by two-sided Welch’s t-test. (I) Schematic representation of the proposed non-canonical role of NR4A1 in cancer progression: effective NR4A1-mediated regulation of IEG expression favors adaptation of cancer cells to chronic replication stress and continued proliferation, whereas genetic or drug-induced suppression of NR4A1 triggers mitotic catastrophe and proliferative failure.