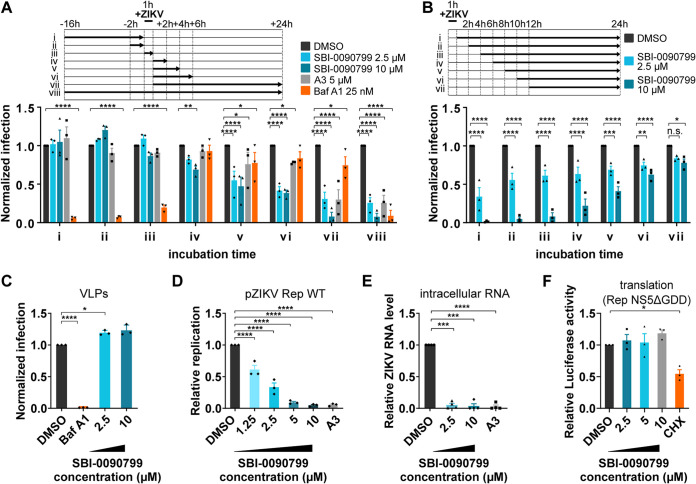
FIG 4.
Mapping the step of the ZIKV replication cycle targeted by SBI-0090799. (A and B) Time-of-addition assays. To synchronize infection, Huh-7.5 cells were infected for 1 h with ZIKV MR766 (MOI of 0.6), and the inoculum was then removed. Cells were also incubated with the indicated compound at the indicated time points. Infection was quantified 24 h postinoculation after fixation and staining for ZIKV envelope protein. Data are normalized to the mean of DMSO-treated wells for each corresponding time point and are presented as mean ± SEM from n = 3 independent experiments. Bafilomycin A1 and compound A3 were used as positive controls at the indicated concentrations. (C) Reporter virus-like particle (VLP) assay. Huh-7.5 cells were infected for 2 h with ZIKV pseudotyped particles harboring a WNV replicon expressing GFP in the presence of the indicated compounds. Inoculum was removed after 2 h, and fresh medium containing the same compounds was added to the wells. GFP-positive cells were quantified 24 h postinoculation. The percentage of infected cells was calculated as the ratio of the number of GFP-positive cells to number of cells stained with DAPI. (D) Huh-7.5 cells were electroporated with RNA encoding the replicon pZIKV Rep WT. The antiviral activity of SBI-0090799 was assessed 24 h postelectroporation by measuring the Renilla luciferase activity as a readout of viral replication. The graph shows the level of replication (mean ± SEM) relative to the DMSO control. Renilla counts were normalized to 4 h posttransfection values, reflecting electroporation efficiency. (E) Huh-7.5 cells were infected with ZIKV MR766 at an MOI of 0.3 in the presence of the indicated compounds. Inoculum was removed after 1 h, and fresh medium supplemented with the compounds was added to the wells. Cells were harvested 24 h postinfection, and the intracellular RNA was isolated and quantified by RT-qPCR. For each condition, ZIKV RNA levels relative to the DMSO-treated wells are shown for n = 3 biological replicates. RPLP0 was used as the housekeeping gene to normalize for input RNA levels. (F) Huh-7.5 cells were electroporated with RNA encoding the ZIKV replicon Rep-NS5ΔGDD. The antiviral activity of SBI-0090799 was assessed 4 h postelectroporation by measuring the Renilla luciferase activity as readout of viral RNA translation. The graph shows the level of luciferase (mean ± SEM) relative to the DMSO control. Cycloheximide (50 μg/ml) is used as a positive control. Results are presented as means ± SEM (n = 3). Two-way (A and B) or one-way (C to F) analysis of variance followed by Tukey’s or Dunnett’s posttest was performed for statistical analysis. Baf A1, bafilomycin A1; CHX, cycloheximide; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.001; n.s., not significant.