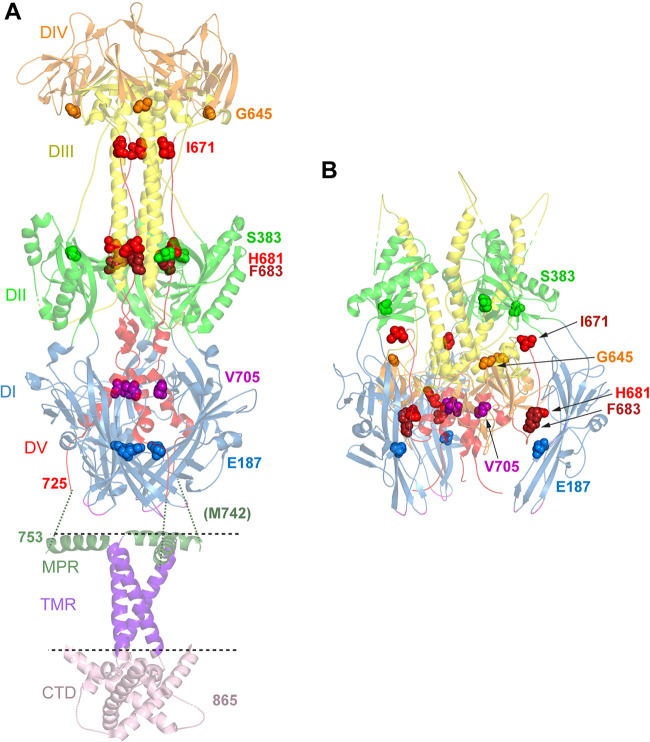
FIG 7.
Location of mutations in revertant viruses. (A) Postfusion structure of full-length HSV-1 gB (PDB accession number 5V2S) (3). Domains (DI to DV) in the ectodomain are colored as described in the legend of Fig. 1. In addition, the membrane proximal region (MPR) (dark green ribbons), transmembrane region (TMR) (purple ribbons), and cytoplasmic tail domain (CTD) (pink ribbons) are shown. The location of the membrane is indicated by dotted lines. The ribbons are partially transparent. The residues mutated in gB3A (I671, H681, and F683) and the residues mutated in the gB3A revertant viruses are shown as space-filled residues. E187 (blue spheres) was mutated in one of the gB3A revertant viruses and lies in domain I near the base of the structure. M742 was mutated in another revertant virus. M742 maps to an unresolved region (green dotted lines) between the C terminus of the ectodomain (position 725) and the N terminus of the MPR (position 753). In a third revertant virus, S383 (green spheres), G645 (orange spheres), V705 (pink spheres), and V880 were mutated. V880 maps downstream of the C terminus of the CTD depicted here. (B) Prefusion model of HSV-1 gB (PDB accession number 6Z9M) (15). Domains and residues in the trimer are colored as described above for panel A. E187 (blue spheres) in domain I contacts domain V of a neighboring protomer, adjacent to F683.