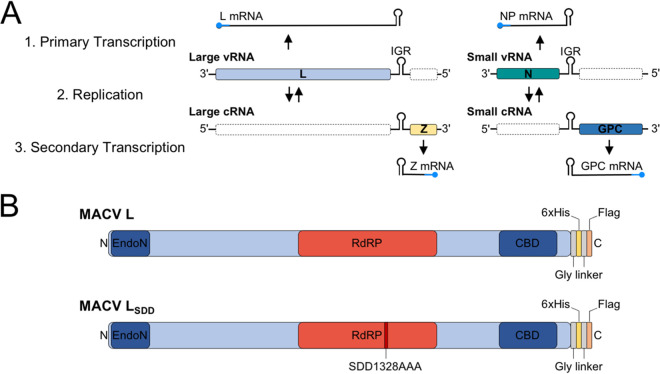
FIG 1.
Arenavirus genome organization and gene expression strategies. (A) Illustration of the mammarenavirus genome and sequential stages of transcription and replication during the infection cycle. The sequential order of primary transcription (L and NP mRNA synthesis), genome replication (cRNA synthesis from vRNA template), and secondary transcription (Z and GPC mRNA synthesis) are listed numerically on the left-hand side. Viral ORFs in the expressed coding orientation are shown on corresponding template RNAs as colored boxes, whereas the antisense ORFs are illustrated as empty white boxes with a dashed outline. 5′ cap structures are shown as rounded-end caps on the mRNAs and are colored blue to indicate nonviral sequences acquired via cap snatching. The intergenic regions (IGRs) of each RNA segment are illustrated between the ORFs as a structured hairpin cartoon. (B) Linear map of the Machupo virus (MACV) L protein. The C-terminal cap-binding domain (CBD) and N-terminal cap-dependent endonuclease (EndoN) are shown in dark blue. The central RNA-dependent RNA polymerase (RdRP) domain is shown in red. The introduced affinity tags described in this study are shown as colored regions at the C terminus of L. The position of the mutated residues (SDD1328AAA) for the catalytically inactive mutant of L (LSDD) is shown in dark red.