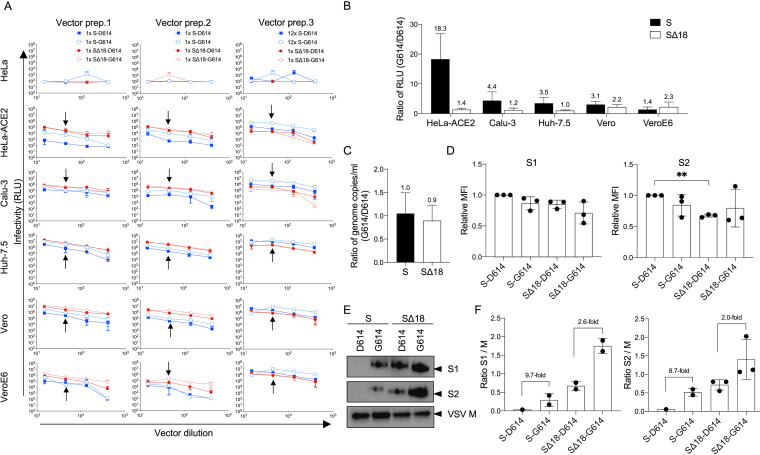
FIG 4.
Impact of D614G mutation is only observed with full-length Spike protein. (A) Indicated cell lines were transduced with G614 or D614 variants of VSV-Luc vectors for both full-length and truncated Spike protein versions. Three preparations of all 4 vectors were produced and tested independently (vector prep. 1 to 3), using the same serial dilutions and volumes of vectors applied to the cells for transduction. For the full-length Spike variants, both unconcentrated (1×) and 12× concentrated vectors were tested. Luciferase activity was measured after 24 h. Means and standard deviations for 3 technical repeats on the same plate are shown. Arrows indicate the luciferase activity (RLU) data points used to compare G614 and D614 variants of specific Spike protein and cell line combinations. (B) Ratio of luciferase activity for the G614:D614 pair combinations indicated, on different cell lines, calculated using the RLU data points arrowed in panel A. Each pair of compared vectors was produced in the same way, and equal volumes were applied in transduction. Means and standard deviations for ratios calculated from each of the three independent vector preparations are shown. (C) Ratio of genomic titers of G614 versus D614 vectors, for both full-length and truncated Spike proteins. Shown are means and standard deviations from equal volumes of 3 independent vector stocks, produced in the same way for each pairwise comparison. (D) Cell surface expression levels of different Spike protein variants on 293T vector-producing cells, measured by flow cytometry using anti-S1 or anti-S2 antibodies at the time of vector harvest. Expression levels are reported as mean fluorescence intensity (MFI) and normalized to the level of S-D614 in each independent experiment. Means and standard deviations from 3 independent experiments are shown. **, P < 0.01, one-way analysis of variance with Tukey’s multiple comparisons. (E) Representative Western blot showing incorporation of Spike proteins into VSV particles, from equal volumes of 100× concentrated vector supernatants, using antibodies against S1 or S2 subunits of Spike or VSV M protein. (F) Comparison of Spike subunit incorporation into vectors, normalized to VSV M. Data are from 2 to 3 independent vector stocks, indicated by individual dots. S1 and S2 subunits were only detected in one stock of S-D614 vectors.