Abstract
Human impact is noticeable around the globe, indicating that a new era might have begun: the Anthropocene. Continuing human activities, including land‐use changes, introduction of non‐native species and rapid climate change, are altering the distributions of countless species, often giving rise to human‐mediated hybridization events. While the interbreeding of different populations or species can have detrimental effects, such as genetic extinction, it can be beneficial in terms of adaptive introgression or an increase in genetic diversity. In this paper, I first review the different mechanisms and outcomes of anthropogenic hybridization based on literature from the last five years (2016–2020). The most common mechanisms leading to the interbreeding of previously isolated taxa include habitat change (51% of the studies) and introduction of non‐native species (34% intentional and 19% unintentional). These human‐induced hybridization events most often result in introgression (80%). The high incidence of genetic exchange between the hybridizing taxa indicates that the application of a genic view of speciation (and introgression) can provide crucial insights on how to address hybridization events in the Anthropocene. This perspective considers the genome as a dynamic collection of genetic loci with distinct evolutionary histories, giving rise to a heterogenous genomic landscape in terms of genetic differentiation and introgression. First, understanding this genomic landscape can lead to a better selection of diagnostic genetic markers to characterize hybrid populations. Second, describing how introgression patterns vary across the genome can help to predict the likelihood of negative processes, such as demographic and genetic swamping, as well as positive outcomes, such as adaptive introgression. It is especially important to not only quantify how much genetic material introgressed, but also what has been exchanged. Third, comparing introgression patterns in pre‐Anthropocene hybridization events with current human‐induced cases might provide novel insights into the likelihood of genetic swamping or species collapse during an anthropogenic hybridization event. However, this comparative approach remains to be tested before it can be applied in practice. Finally, the genic view of introgression can be combined with conservation genomic studies to determine the legal status of hybrids and take appropriate measures to manage anthropogenic hybridization events. The interplay between evolutionary and conservation genomics will result in the constant exchange of ideas between these fields which will not only improve our knowledge on the origin of species, but also how to conserve and protect them.
Keywords: climate change, conservation, genomics, hybrid zones, introgression, speciation
1. INTRODUCTION
Human activities are affecting global environmental processes and patterns, from rapid climate change and radical land‐use changes to the introduction of exotic species, prompting some scientists to declare that we have entered a new era: the Anthropocene. The exact start of the Anthropocene has been a matter of debate and its onset has been connected to different events, such as the rise of deforestation and agriculture (Ruddiman, 2003), the Columbian Exchange of species between the Old World and the New World (Lewis & Maslin, 2015), the Industrial Revolution in the 1800s (Crutzen, 2002), and the population growth and industrialization during the mid‐20th century (Steffen et al., 2015). Based on the increase in the use of certain materials (e.g., aluminum, plastics, concrete), the nuclear fallout of atomic bomb testing, the geochemical signatures of particular compounds (e.g., polyaromatic hydrocarbons and pesticides), and the atmospheric rise in carbon concentrations, Waters et al., (2016) proposed a lower boundary for the Anthropocene during the mid‐20th century. In this review, I will follow this definition to provide a clear distinction between hybridization events that occurred before or during the Anthropocene.
Anthropogenic developments affect the distribution of numerous species, often resulting in secondary contact between previously allopatric taxa. If these taxa are closely related and reproductive isolation is incomplete, hybridization might occur. In this review, I will define hybridization following Arnold (1997), namely the situation in which “two populations of individuals that are distinguishable on the basis of one or more heritable characters overlap spatially and temporally and cross to form viable and at least partially fertile offspring.” These hybrid interactions can have detrimental consequences for the interbreeding populations, such as genetic swamping or extinction by hybridization (Rhymer & Simberloff, 1996; Todesco et al., 2016). This is why conservationists have established guidelines to deal with such cases of human‐induced hybridization (Allendorf et al., 2001). However, hybridization can also provide ecological and evolutionary opportunities, such as the origin of new hybrid species (Mallet, 2007; Ottenburghs, 2018) or the exchange of adaptive genetic variation (Arnold & Kunte, 2017; Hedrick, 2013). It is thus important to determine the balance between these potential detrimental and beneficial consequences when devising effective conservation strategies.
In the following sections, I first review the literature on anthropogenic hybridization from the last five years (2016–2020) to identify the most common mechanisms and outcomes of human‐mediated hybridization events (see Table 1, extending the search strategy from Grabenstein & Taylor, 2018). Next, I introduce the “genic view of speciation,” a concept that has shaped recent research agendas in speciation genomics (Campbell et al., 2018; Ravinet et al., 2017; Wu, 2001). This viewpoint focuses on the heterogenous nature of genetic differentiation and introgression across the genome. The insight that different genomic regions tell different evolutionary stories needs to be taken into account when dealing with anthropogenic hybridization events. Specifically, it has important consequences for (1) the development of molecular markers, (2) the quantification of (adaptive) introgression patterns, and (3) the legal status of hybrids.
TABLE 1.
Overview of studies on anthropogenic hybridization events published in the last five years (2016–2020)
| Species | Location | Mechanism | Outcome | Molecular markers | Reference |
|---|---|---|---|---|---|
| Plants | |||||
|
Centaurea seridis Centaurea aspera |
Spain | Habitat Change |
F1 hybrids (triploids) |
NA | Garmendia et al., (2018) |
| Eucalyptus tetrapleura | Australia | Habitat Change | Introgression | RADseq | Rutherford et al., (2018) |
| Odontarrhena spp. | Albania |
Habitat Change |
Introgression | AFLP‐fingerprinting | Coppi et al., (2020) |
|
Phragmites australis Phragmites mauritanius |
Southern Africa | Habitat Change | Introgression |
Chloroplast DNA Microsatellites |
Canavan et al., (2018) |
| Nevada, USA | Introduction | F1 hybrids | Saltonstall et al., (2016) | ||
|
Quercus durata Quercus berberidifolia |
California, USA |
Habitat Change (fire frequency) |
Introgression | Microsatellites | Ortego et al., (2017) |
|
Rhododendron ferrugineum Rhododendron hirsutum |
Italy | Habitat Change | Introgression |
Chloroplast DNA Microsatellites |
Bruni et al., (2016) |
|
Taraxacum calanthodium Taraxacum lugubre |
China | Pollination by introduced bees | Introgression | Microsatellites | Peng et al., (2018) |
| Insects | |||||
|
Bactrocera tryoni Bactrocera aquilonis |
Australia |
Habitat Change (horticulture) |
Introgression | RADseq | Popa‐Báez et al., (2020) |
|
Helicoverpa armigera Helicoverpa zea |
Brazil |
Introduction (invasive species) |
Introgression * (adaptive) |
Whole Genome | Valencia‐Montoya et al., (2020) |
|
Phaulacridium marginale Phaulacridium otagoense |
New Zealand |
Habitat Change (deforestation) |
Introgression | Mitochondrial and nuclear loci | Sivyer et al., (2018) |
| Amphibians | |||||
|
Bufo woodhousii Bufo microscaphus |
Southwest USA | Habitat Change | Introgression | Microsatellites | Wooten et al., (2019) |
|
Hyperolius thomensis Hyperolius molleri |
Sao Tomé Island |
Habitat Change (deforestation) |
Introgression |
mtDNA RADseq |
Bell and Irian (2019) |
|
Lissotriton vulgaris meridionalis Lissotriton vulgaris vulgaris |
Italy | Introduction |
F1 hybrids |
Mitochondrial and nuclear loci | Dubey et al., (2019) |
| Pelophylax spp. | Switzerland | Introduction | F1 hybrids |
mtDNA Microsatellites |
Dufresnes et al., (2018) |
| Rana pipiens | Southern USA | Introduction | Introgression |
mtDNA Microsatellites |
O’Donnell et al., (2017) |
|
Triturus cristatus Triturus carnifex |
The Netherlands | Introduction | Introgression | Mitochondrial and nuclear loci | Wielstra et al., (2016) |
| Reptiles | |||||
| Pelodiscus spp. | China |
Introduction (farmed turtles) |
Introgression | Mitochondrial and nuclear loci | Gong et al., (2018) |
|
Sternotherus depressus Sternotherus peltifer |
Alabama, USA | Habitat Change |
Introgression (unidirectional) |
RADseq | Scott et al., (2019) |
| Fish | |||||
|
Archosargus probatocephalus Archosargus rhomboidalis |
Florida, USA | Habitat Change | F1 hybrids |
mtDNA & nDNA microsatellites |
Seyoum et al., (2020) |
|
Alosa alosa Alosa fallax |
France | Habitat Change | Introgression | Mitochondrial and nuclear loci | Taillebois et al., (2020) |
| Alosa pseudoharengu | Connecticut, USA | Introduction | Introgression | RADseq | Reid et al., (2020) |
|
Catostomus discobolus Catostomus ardens |
USA | Habitat Change | F1 hybrids | Mitochondrial and nuclear loci | Bangs et al., (2017) |
|
Cobitis magnostriata Cobitis minamorii oumiensis |
Japan | Habitat Change |
Reproductive Interference |
mtDNA | Morii et al., (2018) |
|
Colpichthys regis Colpichthys hubbsi |
California, USA | Habitat Change |
Introgression (unidirectional) |
mtDNA Microsatellites |
Lau and Jacobs (2017) |
|
Coregonus lavaretus (wild and stocked) |
Finland |
Introduction & Habitat Change |
Introgression | Microsatellites | Huuskonen et al., (2017) |
| Coregonus spp. | Switzerland |
Habitat Change (eutrophication) |
Introgression | RADseq | Feulner and Seehausen (2019) |
|
Cottus sp. Cottus cognatus |
Canada | Habitat Change | Introgression |
mtDNA Microsatellites |
Rudolfsen et al., (2019) |
|
Etheostoma osburni Etheostoma variatum |
West Virginia, USA |
Habitat Change |
Introgression | Microsatellites | Gibson et al., (2019) |
|
Fundulus grandis Fundulus heteroclitus |
Mexico | Habitat Change |
Introgression * (adaptive) |
Whole Genome | Oziolor et al., (2019) |
|
Gila cypha Gila robusta |
Colorado, USA | Habitat Change | Introgression | RADseq | Chafin et al., (2019) |
|
Macquaria ambigua (wild and stocked) |
Australia | Introduction | Introgression | RADseq | Beheregaray et al., (2017) |
| Micropterus spp. | Southern USA | Introduction | Introgression | Mitochondrial and nuclear loci | Bangs et al., (2018) |
|
Oncorhynchus chrysogaster Oncorhynchus mykiss |
Mexico |
Introduction (aquaculture) |
Introgression | RADseq | Escalante et al., (2020) |
|
Oncorhynchus clarkii Oncorhynchus mykiss |
USA | Introduction | Introgression | Allozymes, SNPs and microsatellites |
Muhlfeld et al., (2017) |
| Oncorhynchus tshawytscha | Washington, USA | Habitat Change | F1 hybrids | SNP markers | Fraser et al., (2020) |
| Oreochromis niloticus | Ethiopia | Introduction | Introgression | Microsatellites | Tibihika et al., (2020) |
|
Parachondrostoma toxostoma Chondrostoma nasus |
France | Habitat Change | Introgression |
mtDNA Microsatellites |
Guivier et al., (2019) |
|
Salvelinus alpinus Salvelinus fontinalis |
Sweden | Introduction | Introgression |
mtDNA Microsatellites |
Faulks and Östman (2016) |
|
Salvelinus fontinalis (wild and stocked) |
New York, USA | Introduction | Introgression | Microsatellites | Bruce et al., (2020) |
|
Salvelinus fontinalis Salvelinus confluentus |
Oregon, USA |
Introduction & Habitat Change |
F1 hybrids (morphology) |
NA | Howell (2018) |
|
Sander vitreus Sander Canadensis |
Canada |
Introduction & Habitat Change |
Introgression | RADseq | Graham et al., (2020) |
|
Tropheus moorii (two lineages) |
Lake Tanganyika |
Habitat Change (lake levels) |
Introgression | mtDNA, AFLP, and Microsatellites | Sefc et al., (2017) |
| Birds | |||||
|
Alectoris rufa Alectoris chukar |
Italy |
Introduction (hunting) |
Introgression | mtDNA | Forcina et al., (2020) |
|
Francolinus francolinus (subspecies) |
Pakistan | Introduction | Introgression |
mtDNA Microsatellites |
Forcina et al., (2018) |
|
Mycteria cinerea Mycteria leucocephala |
Singapore |
Introduction (captive hybrids) |
Introgression | RADseq | Baveja et al., (2019) |
|
Vermivora chrystoptera Vermivora cyanoptera |
Canada | Habitat Change | F1 Hybrids | mtDNA | Moulton et al., (2017) |
| Mammals | |||||
|
Aepyceros melampus petersi Aepyceros melampus melampus |
Namibia & South Africa | Introduction | Introgression | Microsatellites | Miller et al., (2020) |
| Callithrix spp. | Brazil | Introduction | Introgression | NA | Malukiewicz (2019) |
|
Canis lupus Canis lupus familiaris |
Italy |
Introduction (domestic dogs) |
Introgression | Nuclear loci | Salvatori et al., (2019) |
|
Cervus elaphus Cervus nippon |
Scotland | Introduction | Introgression | SNP genotyping | McFarlane et al., (2020) |
| Cervus elaphus | Spain & Portugal |
Introduction (non‐native deer) |
Introgression |
mtDNA Microsatellites |
Queirós et al., (2020) |
|
Damaliscus pygargus pygargus Damaliscus pygargus phillipsi |
South Africa | Introduction | F1 hybrids | Microsatellites | Van Wyk et al., (2017) |
|
Felis silvestris Felis catus |
Europe |
Introduction (domestic cats) |
Introgression |
SNP genotyping Microsatellites |
Tiesmeyer et al., (2020) |
|
Gazella bennettii Gazella subgutturosa |
Iran | Introduction | F1 Hybrids | Mitochondrial and nuclear loci | Fadakar et al., (2020) |
|
Sus scrofa (wild and domesticated) |
Japan |
Introduction (escaped animals) |
Introgression | mtDNA | Anderson et al., (2019) |
| Other taxa | |||||
|
Daphnia pulex (different lineages) |
Canada | Habitat Change | Introgression |
mtDNA Microsatellites |
Millette et al., (2020) |
|
Daphnia longispina Daphnia galeata |
Switzerland |
Habitat Change (eutrophication) |
Introgression |
mtDNA Microsatellites |
Alric et al., (2016) |
|
Pomacea canaliculata Pomacea maculata |
Brazil & Uruguay | Introduction | Introgression | Nuclear markers | Glasheen et al., (2020) |
These studies were found in a systematic literature search of Google Scholar and the Web of Science, using the combined search terms: ‘anthropogenic AND disturb* AND hybrid*’OR ‘habitat AND chang* AND hybrid*’ OR ‘human AND chang* AND hybrid*’ OR ‘environment* AND chang* AND hybrid*’ NOT climate NOT introduc* NOT zone* (following Grabenstein & Taylor).
2. MECHANISMS OF ANTHROPOGENIC HYBRIDIZATION
There are three main mechanisms of anthropogenic hybridization (Figure 1), namely human‐assisted translocations, habitat modifications, and climate change (Crispo et al., 2011; Grabenstein & Taylor, 2018). Translocations or introductions of certain taxa can be intentional, such as in genetic rescue programs. This strategy is used to restore genetic diversity and reduce the extinction risk of small, isolated, and often inbred populations. About 90 percent of these genetic rescue attempts have been successful, indicating that human‐mediated hybridization can be an important tool in conservation (Frankham, 2015). In this review, however, I focus on species translocations and introductions that are unrelated to conservation efforts. My literature search uncovered 31 papers (out of 59 studies, 53%) that involved the introduction of non‐native species (Figure 2a). Some of these cases involved intentional movement of organisms, such as the release of game‐farm birds for hunting purposes (Forcina et al., 2020), the stocking of fish populations with captive‐bred animals (Beheregaray et al., 2017; Bruce et al., 2020), or the translocation of large mammals between African game reserves (Grobler et al., 2018; Miller et al., 2020; van Wyk et al., 2019). Other human‐assisted translocations were unintentional, such as the transport of aquatic organisms in ship hulls (Oziolor et al., 2019) or plant seeds by cargo or passenger transport, sometimes resulting in cryptic invasions and hybridization events (Morais & Reichard, 2018). In addition, domestic animals that escape from captivity often interbreed with their wild relatives (Anderson et al., 2019; Salvatori et al., 2019; Tiesmeyer et al., 2020). Species translocations or introductions—both intentional and unintentional—leading to hybridization events have thus been documented in a variety of taxa and are expected to increase as humans continue to move species across the globe (Crispo et al., 2011).
FIGURE 1.
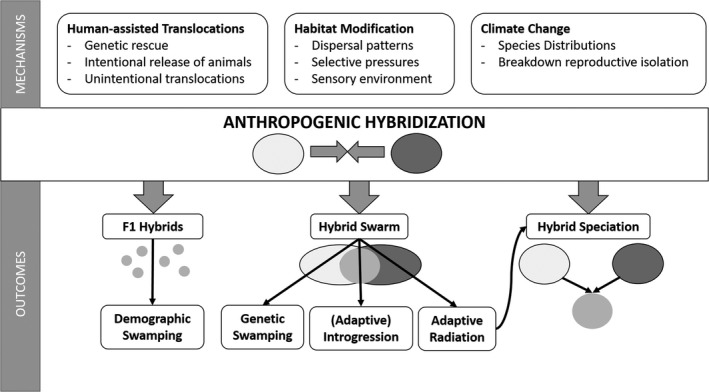
Mechanisms and outcomes of anthropogenic hybridization. Mechanisms that can result in hybridization include human‐assisted translocations (both intentional and unintentional), habitat modification and climate change. Anthropogenic hybridization events can result in the formation of first‐generation (F1) hybrids, a hybrid swarm or a hybrid species. If the production of F1 hybrids interferes with the reproductive output of the parental species, demographic swamping can occur. A hybrid swarm can lead to (adaptive) introgression, genetic swamping or provide the raw material for an adaptive radiation (potentially including the origin of a hybrid species)
FIGURE 2.
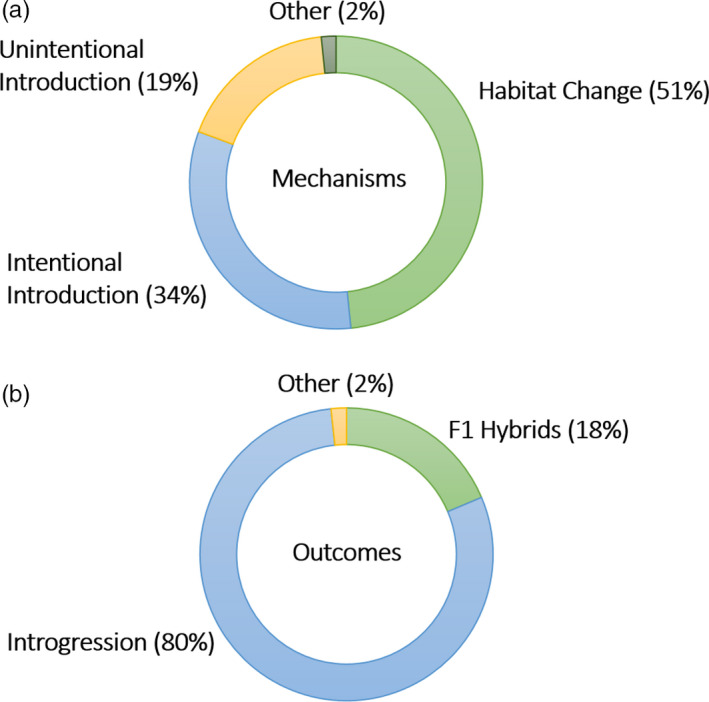
The most common mechanisms and outcomes of anthropogenic hybridization, based on a literature review of the last five years (2016–2020). Habitat change is the most prevalent mechanism behind human‐induced hybridization events, followed by intentional and unintentional translocations of organisms. The vast majority of anthropogenic hybridization events results in introgression
Anthropogenic hybridization can also be the outcome of habitat modifications (Grabenstein & Taylor, 2018). Edgar Anderson (1948) coined the phrase “hybridization of the habitat” to indicate that human disturbances of the environment can lead to the production of hybrid offspring. Indeed, 30 studies (51%) attributed hybridization events to human‐induced habitat changes (Figure 2a). In several plant species, the rate of hybridization increased with the level of disturbance in the area, such as the concentration of heavy metals (Coppi et al., 2020) or the frequency of wildfires (Ortego et al., 2017). Hybridization can also be the outcome of homogenization of the habitat or the construction of landscape features that facilitate dispersal (e.g., roads and verges), culminating in contact between previously isolated populations (Bangs et al., 2017; Carantón‐Ayala et al., 2018; van Hengstum et al., 2012). For instance, deforestation has led to hybridization between certain insect (Sivyer et al., 2018) and amphibian populations (Bell & Irian, 2019). Moreover, habitat disturbances can affect selective pressures, allowing hybrids to thrive in new environments that are not accessible for the parental populations (Arnold et al., 2012; Arnold & Martin, 2010). For example, hybrids between cave salamanders (Hydromantes ambrosii and Hydromantes italicus) with transgressive phenotypes could expand into more harsh environments with higher food availability (Ficetola et al., 2019). Human‐mediated actions can also influence the sensory environment in a number of ways, such as pollutants that alter chemical signaling (Smadja & Butlin, 2009) or anthropogenic noise that interferes with auditory communication (Slabbekoorn et al., 2010; Slabbekoorn & Ripmeester, 2008). A well‐studied example concerns the effect of increased lake eutrophication on species recognition in fish (Alexander et al., 2017; Vonlanthen et al., 2012). Eutrophication decreases water clarity, leading to a breakdown of prezygotic barriers because individuals cannot discriminate between conspecifics and heterospecifics in the turbid waters (Seehausen et al., 1997). As human‐mediated habitat modifications continue to change the environmental and sensory conditions for numerous taxa, more hybridization events are expected to arise in the near future (Crispo et al., 2011; Grabenstein & Taylor, 2018).
Climate change can be seen as a special case of habitat change and can affect hybridization dynamics in a myriad of ways (Chunco, 2014). Both latitudinal and altitudinal range shifts are expected to occur, leading to secondary contact between previously isolated species (Larson et al., 2019; Parmesan, 2006; Taylor et al., 2015). Such distributional shifts and consequent hybridization events have already been documented, for instance between polar bears (Ursus maritimus) and grizzly bears (U. arctos) in the Arctic (Kelly et al., 2010) and between species of Glaucomys flying squirrels (Garroway et al., 2010), and will be observed more often in the future (Chunco, 2014; Taylor et al., 2015). Apart from distributional changes, climate change might also result in the breakdown of reproductive isolation mechanisms between sympatric species, such as the disappearance of temporal isolation due to phenological changes (Vallejo‐Marín & Hiscock, 2016). Climate change will thus alter population dynamics in time and space, culminating in more hybridization events.
3. OUTCOMES OF ANTHROPOGENIC HYBRIDIZATION
Regardless of the underlying mechanism, anthropogenic hybridization can have several outcomes, which are mainly determined by the level of genetic divergence and the nature of reproductive isolation between the interacting species (Figure 1). In some cases, the hybridization event is limited to first‐generation (F1) hybrids (11 studies, 19%, Figure 2b), such as the production of triploid hybrids between diploid and tetraploid Centaurea species in Spain (Garmendia et al., 2018) or the occasional hybrid between subspecies of the smooth newt (Lissotriton vulgaris) in Italy (Dubey et al., 2019). The absence of second‐generation hybrids or backcrosses might be due to the lower fitness of F1 hybrids (e.g., decreased fertility) or the lower sensitivity of certain molecular markers that fail to identify later‐generation hybrids. Another possible outcome of human‐induced hybridization is a hybrid swarm: a population of fertile hybrids that survived past the first hybrid generation, followed by interbreeding between hybrid individuals and backcrossing with their parental populations. Hybrid swarms generally form within a hybrid zone, an area where two populations overlap spatially and temporally and produce viable and at least partially fertile offspring within a restricted area (e.g., Carney et al., 2000; Fisher et al., 2006). In the majority of studies (47 studies, 80%), the production of hybrids resulted in the exchange of genetic material through backcrossing with parental species (i.e., introgression, Figure 2b). On a longer timeframe, the fate of these hybrid swarms is difficult to predict. The mixing of different genetic lineages might lead to the extinction of particular lineages through species collapse (Zhang et al., 2019) or it might provide the raw material for an adaptive radiation (Marques et al., 2019; Seehausen, 2004). In theory, hybrid species can emerge from hybrid swarms, but this speciation process is typically too slow to observe on a short timescale (Lamichhaney et al., 2018; Ottenburghs, 2018; Seehausen, 2004). There are, however, some exceptions, such as the Oxford ragwort (Senecio squalidus) that originated from the interbreeding of two Italian species (S. aethnensis and S. chrysanthemifolius) in British gardens at the turn of the 18th century (Nevado et al., 2020).
On a population level, different combinations of hybrid fitness and hybridization rate can lead to drastically different outcomes in an anthropogenic hybridization event. First, if hybrid fitness is comparable to the fitness of parents (i.e., outbreeding depression is low) and hybrids show higher population growth rates than the parental taxa, genetic swamping might occur in which the parental taxa are replaced by hybrids (Rhymer & Simberloff, 1996). This might be occurring in West Virginia (USA) where hybrids between candy darters (Etheostoma osburni) and variegate darters (E. variatum) are replacing the now endangered candy darter (Gibson et al., 2019). A special case of potential genetic swamping concerns the release of actively managed species, such as fish stocks and game birds (Randi, 2008). These artificially selected organisms often differ genetically from their wild conspecifics which can result in outbreeding depression when they interbreed (Muhlfeld et al., 2009). Moreover, the strong selection pressures in captivity might lead to low genetic diversity and inbreeding depression (Willoughby et al., 2015). Hybridization with these maladapted individuals can lower the average fitness of wild populations. Although most studies documented introgression and warn for the possibility of genetic swamping, few studies directly quantified the likelihood of genetic swamping in their study system. This important knowledge gap should be addressed with more detailed analyses, complemented with modeling studies, to determine crucial tipping points in introgression levels that can lead to genetic swamping.
If hybrid fitness is strongly reduced compared to the parental taxa (i.e., outbreeding depression is high) and hybridization rates are high, parental taxa will waste reproductive effort when hybridizing. This situation can lead to demographic swamping where parental taxa decline due to decreased population growth rates (Wolf et al., 2001). This situation has been described for the Australian native variable groundsel (Senecio pinnatifolius) and the invasive Madagascar ragwort (S. madagascariensis). Hybridization rates between these species are very high, but hybrids are not viable. As the invasive plant increases in abundance, the native plant continues to waste reproductive effort and will start declining in numbers (Prentis et al., 2007). Another example concerns reproductive interference between two spined loaches (Cobitis magnostriata and C. minamorii oumiensis) that could lead to the decline of the latter species (Morii et al., 2018). Between the extremes of genetic and demographic swamping, there is a continuum of introgression patterns that are neutral or benefit one or both hybridizing taxa.
4. THE GENIC VIEW OF SPECIATION AND INTROGRESSION
This overview of the different mechanisms and outcomes of anthropogenic hybridization highlights the difficulty in predicting the future developments of hybrid interactions and devising appropriate conservation measures. Recent developments in genomic tools have led to more accurate detection of hybrids and the quantification of introgression patterns (McFarlane & Pemberton, 2019), but an overarching framework is needed to interpret these findings and translate them to successful conservation strategies. Here, speciation genomics and the study of pre‐Anthropocene hybridization events can provide additional insights (Campbell et al., 2018; Taylor et al., 2015). In particular, the genic view of speciation has drastically changed the way evolutionary biologists study the process of speciation (Bazykin, 1969; Key, 1968; Wu, 2001) and this perspective can be applied to several conservation genomic questions.
Speciation research has long been dominated by the Biological Species Concept (BSC) which focuses on reproductive isolation between diverging lineages (Mayr, 1963). This concept assumed that species differentiation is controlled by a large number of genetic loci and that the whole genome functions as an integrated and cohesive genetic unit. Hybridization and consequent genetic exchange were thought to destroy this integrity and break up “co‐adapted gene complexes.” This perspective is still widely followed by conservationists that describe introgressive hybridization as “genetic erosion” (Chafin et al., 2019) or “genetic pollution” (Wielstra et al., 2016). However, genomic studies have overturned the idea of the genome as a cohesive genetic unit that can be destroyed by hybridization. Instead, the genome can better be regarded as a dynamic collection of genetic loci with separate, but entangled evolutionary histories. Moreover, reproductive isolation is often controlled by epistatic interactions between a few genetic loci (Ravinet et al., 2017; Wu, 2001).
This new perspective is nicely illustrated by recent studies that showed how genetic differentiation between diverging lineages is heterogeneously distributed across the genome, often concentrated in a few genomic regions, so‐called “islands of differentiation” (Wolf & Ellegren, 2017). The genomic islands may harbor loci that contribute to reproductive isolation, and these so‐called barrier loci are thus less likely to introgress compared to neutral loci. Consequently, barrier loci and closely linked genomic regions will diverge while introgression homogenizes the rest of the genome (Ravinet et al., 2017). An alternative explanation for the origin of genomic islands concerns linked selection, which comprises two processes: background selection and genetic hitchhiking (Burri, 2017). Background selection refers to purifying selection against recurring deleterious mutations, while genetic hitchhiking occurs when positive selection on a variant result in the selection for the genomic region in which this advantageous variant resides. As the advantageous variant increases in frequency, the loci linked to this variant hitchhike along (Sendell‐Price et al., 2020). Regardless of the underlying process—barrier loci or linked selection—the end result is a heterogenous genomic landscape of differentiation, which affects several aspects of conservation genomics, including (1) the development of molecular markers, (2) the study of introgression, and (3) the legal status of hybrids.
5. DEVELOPMENT OF MOLECULAR MARKERS
A first step in the assessment of an anthropogenic hybridization event involves determining the composition of the hybrid population in terms of parental individuals, first‐generation hybrids (F1), second‐generation hybrids (F2), backcrosses, etc. For example, a mixture of different generational hybrids indicates the formation of a hybrid swarm, whereas the absence of backcrosses and second‐generation hybrids suggests selection against hybrids. Most conservation genetic studies use a set of variable genetic markers (e.g., microsatellites) to quantify the genetic make‐up of different individuals using software packages such as STRUCTURE (Pritchard et al., 2000) or ADMIXTURE (Alexander et al., 2009). Next, this genetic make‐up can be compared with simulated data to test the power of discriminating between different levels of admixture (Anderson, 2008). The resulting composition of the hybrid population provides the basis for subsequent analyses and potential conservation measures.
The use of a few genetic markers works well to identify early‐generation hybrids and backcrosses, but can run into several issues (McFarlane & Pemberton, 2019). First, the determination of a threshold to discriminate between hybrid and parental species can be problematic. Population assignment algorithms calculate an admixture score (Q) for individuals where a score of 0 or 1 indicates a purebred individual from one of the parental populations. However, due to errors in genotyping or the presence of nondiagnostic markers, most individuals will have a score that is not exactly 0 or 1. But how does one discriminate between these errors and actual hybrids (which have a score between 0 and 1)? Generally, a threshold is used to delimit the parental classes, but this threshold varies between studies, ranging from 0.8 (Schulte et al., 2012) to 0.99 (Galaverni et al., 2017), and the choice of the threshold depends on the number of hybrid classes one recognizes. Using a threshold of 0.8 would, for instance, assign some later‐generation backcrosses to a parental class. A second issue of using few genetic makers for hybrid detection concerns homozygous loci. By chance, several generations of backcrossing can result in homozygous genetic markers for some individuals, which will consequently not be recognized as hybrids or backcrosses (Boecklen & Howard, 1997).
These two issues—setting a realistic admixture threshold and the presence of homozygous markers—can often be solved by adding more markers. Indeed, genomic studies can more confidently identify hybrids and later‐generation backcrosses, leading to more sound conclusions about the hybridization dynamics (Lemopoulos et al., 2019; McFarlane et al., 2020; McFarlane & Pemberton, 2019). For instance, analyses based on microsatellite markers suggested that hybridization between mallards (Anas platyrhynchos) and American black ducks (Anas rubripes) might lead to genetic extinction of the latter species (Mank et al., 2004). However, genomic studies of this system revealed little gene flow between the species, indicating that hybridization is not threatening the genetic integrity of the American black duck (Lavretsky et al., 2019, 2020). Similarly, McFarlane et al., (2020) investigated the sensitivity of microsatellites and RADseq (restriction site‐associated DNA sequencing) to discriminate between different hybrid classes of red deer (Cervus elaphus) and Japanese sika (C. nippon). The RADseq data were able to identify more advanced backcrosses compared to the microsatellites, leading to a more fine‐grained picture of introgression dynamics between these species.
The discrepancy between microsatellite markers and genomic data can partly be explained by the underlying genomic landscape of differentiation. The random selection of a few genetic markers might result in a marker set that only captures the undifferentiated section of the genome, missing the genomic islands of differentiation. Genomic sequencing methods, such as RADseq, cover a larger proportion of the genome compared to microsatellites and can be used to develop diagnostic markers. For instance, Taillebois et al., (2020) developed 77 species‐specific SNPs (single nucleotide polymorphisms) that could detect hybrids and backcrosses between allis shad (Alosa alosa) and twaite shad (A. fallax) up to the third generation. Similar marker sets and sequencing protocols have been developed for other study systems (Feulner & Seehausen, 2019; Vaux et al., 2021; Wielstra et al., 2016) and show that conservationists do not always need whole genome sequencing data to reconstruct the entire genomic landscape. This conclusion is further supported by evolutionary studies that used less powerful sequencing methods (e.g., RADseq or ultraconserved elements) to explore the genomic landscape of differentiation (Battey, 2019; Bourgeois et al., 2020; Oswald et al., 2019; Plomion et al., 2018). There are, however, certain situations where these methods do not provide the necessary resolution to investigate genetic differentiation across the genome, such as polyploids (Bourke et al., 2018; Clark et al., 2019), large genome sizes (Lowry et al., 2016), or populations with large‐scale demographic changes (Arnold et al., 2013). It is thus important to be conscious about the potential biases of reduced representation methods, such as RADseq (reviewed in Andrews et al., 2016). Nonetheless, being aware of the heterogenous nature of the underlying genomic landscape will already lead to a more conscious selection of diagnostic markers, even if not using whole genome sequencing.
6. PATTERNS OF ANTHROPOGENIC INTROGRESSION
Hybridization often results in introgression: the exchange of genetic material between populations through backcrossing (Ottenburghs, Kraus, et al., 2017; Ottenburghs, Megens, et al., 2017; Taylor & Larson, 2019). Introgression can have detrimental effects, such as loss of genetic integrity, speciation reversal (Seehausen et al., 2008), and the genetic extinction of certain taxa (Rhymer & Simberloff, 1996). However, introgression can be beneficial by facilitating the exchange of adaptive loci and increasing genetic diversity (Arnold & Kunte, 2017; Hedrick, 2013), possibly improving the adaptive potential of a population or species (Funk et al., 2019; Milot et al., 2020). Indeed, several authors have suggested that hybridization can be used as an effective conservation measure (Chan et al., 2019; vonHoldt et al., 2018). From a conservation point of view, we are thus faced with a difficult dilemma when assessing a human‐induced hybridization event: should we prevent potential genetic extinction with conservation measures (e.g., culling hybrids) or should we not intervene to provide the opportunity for adaptive introgression and an increase in genetic diversity?
The positive effects of introgressive hybridization have been well‐documented in several genetic rescue programs (Frankham, 2015; Whiteley et al., 2015), but does it also occur in unintentional anthropogenic hybridization events? A literature survey reported that the majority of human‐induced hybridization events led to an increased extinction risk of the parental species (Todesco et al., 2016), indicating that introgression was mainly detrimental. The discrepancy between the success of intentional genetic rescue programs and the detrimental effects of unintentional anthropogenic hybridization events can be partly explained by the genetic divergence between the hybridizing taxa (Whiteley et al., 2015). Genetic rescue involves the carefully planned supplementation of a small inbred population with individuals from other populations that belong to the same (Heber et al., 2013; Miller et al., 2012) or a closely related subspecies (Harrisson et al., 2016; Pimm et al., 2006) in order to restore genetic diversity. Because the genetic distance between the local and supplemented individuals is low, it is unlikely that the resulting hybrids will suffer from outbreeding depression (i.e., decreased fitness of hybrids relative to their parents). In fact, hybrids in genetic rescue programs often exhibit a temporary increase in fitness due to the masking of rare deleterious alleles (Pickup et al., 2013; Weeks et al., 2017). In contrast to genetic rescue interventions, anthropogenic hybridization events often occur between more distantly related taxa, increasing the likelihood of outbreeding depression due to negative epistatic interactions between divergent loci (Edmands, 1999). Indeed, the majority of studies in my literature search (43 out of 59 studies, 73%) involved different species.
Speciation genomics—particularly the study of pre‐Anthropocene hybrid zones—has shown that introgression varies across the genome (Payseur, 2010). Genetic loci can roughly be divided into three categories: (1) neutral loci that flow freely between taxa, (2) deleterious loci that contribute to reduced fitness in hybrids and inhibit introgression, and (3) beneficial loci that confer an adaptive advantage and increase in frequency following introgression. Admixed genomes can be seen as a mosaic of these three categories, shaped by genetic drift, recombination, and selection (Runemark et al., 2019). This genic view of introgression illustrates that the dichotomy between the deleterious and beneficial effects of introgression on the population level becomes more nuanced on the genomic level (Wu, 2001). Both effects can be acting simultaneously within a collection of genomes: some genomic regions will be homogenized by (adaptive) introgression, while other regions will remain species‐specific due to strong negative selection against introgressed loci.
The majority of conservation genetic studies only quantified the level of introgression between interacting populations, but they did not assess which genomic loci are being exchanged or not. Genomic regions that contribute to reproductive isolation, either because they contain barrier loci or because they are involved in local adaptation, do not introgress and will thus preserve species integrity, even in the face of high levels of hybridization. A study only quantifying introgression might thus warn for genetic swamping or species collapse, even though species‐specific loci will prevent this from happening. For instance, extensive introgression between taiga bean goose (Anser fabalis fabalis) and tundra bean goose (A. f. serrirostris) resulted in a largely homogeneous genomic landscape, but a few genomic islands of differentiation seem to prevent these taxa from merging (Ottenburghs et al., 2020). However, if reproductive isolation mechanisms break down completely, such as the loss of species recognition between fish species due to eutrophication (Vonlanthen et al., 2012), the genomic barriers preventing species from merging have been broken and the underlying loci can also introgress. Hence, the threat of genetic swamping might increase and conservation measures will be warranted. It is thus important to investigate the functional role of introgressed regions. If the genes at the exchanged genomic locations are important in adaptation to rapidly changing environments (e.g., immune genes), a local increase in genetic diversity can provide the necessary adaptive potential to deal with these challenges (Chan et al., 2019; Derry et al., 2019). The development of more powerful techniques to detect introgression (Hibbins & Hahn, 2021) and the increasing completeness and better annotation of genome assemblies (Peona et al., 2018) suggest that these types of analyses will be possible for nonmodel organisms in the near future.
The genic view of introgression also becomes apparent in studies that documented the exchange of adaptive traits between taxa. In most cases, these traits can be traced back to one or a few genetic loci, such as polymorphisms in the gene vkorc1 that causes resistance to rodenticides and introgressed between European and Algerian mouse populations (Song et al., 2011). Similarly, adaptive changes in coat color have been linked to particular introgressed loci in wolves (Anderson et al., 2009) and snowshoe hares (Jones et al., 2018). Examples of adaptive introgression in plants include the transfer of herbivore resistance traits between Helianthus sunflowers (Whitney et al., 2006) and regulatory genes for certain ecological traits between Senecio plants (Kim et al., 2008). Two recent studies on anthropogenic hybridization documented adaptive introgression. Oziolor et al., (2019) showed that Gulf killifish (Fundulus grandis) adapted to high levels of water pollution through introgressive hybridization with the non‐native Atlantic killifish (F. heteroclitus). And a study on hybridization between the local moth species Helicoverpa zea and the invasive H. armigera revealed that an insecticide‐resistant locus introgressed into the local species and increased in frequency (Valencia‐Montoya et al., 2020). Interestingly, both studies relied on whole genome sequencing data, suggesting that it is difficult to detect adaptive introgression with less powerful sequencing methods. However, the methods for detecting adaptive introgression are improving (Moest et al., 2020; Setter et al., 2020; Zhang et al., 2020) and it will only be a matter of time before these methods can be applied more widely, as exemplified by the increasing availability of test statistics to detect introgression (Hibbins & Hahn, 2021). However, not intervening in anthropogenic hybridization events to provide the opportunity for adaptive introgression is generally not advisable and requires a thorough understanding of the study system (Allendorf et al., 2001). In summary, it is not only important to quantify how much genetic material introgressed, but also what has been exchanged.
7. INSIGHTS FROM PRE‐ANTHROPOCENE HYBRIDIZATION EVENTS?
Several anthropogenic hybridization events occur between species that have previously not overlapped in range. For example, hybridization between Corbicula clams in the European river Rhine probably started with the introduction of American lineages through cargo ships (Pfenninger et al., 2002). When two previously allopatric taxa interact for the first time prezygotic isolation mechanisms are often not well developed, resulting in high level of interbreeding and possibly introgression. Indeed, reproductive isolation is often weaker between allopatric taxa compared to sympatric taxa (Coyne & Orr, 1989; Presgraves, 2002), probably because reproductive isolation between sympatric taxa has been strengthened by reinforcement (Calabrese & Pfennig, 2020; Coughlan & Matute, 2020). Alternatively, some hybridizing taxa might have a history of hybridization with previous periods of secondary contact. For instance, polar bears and grizzly bears, that are currently interbreeding in the Arctic due to recent climate change (Kelly et al., 2010), have an extensive history of pre‐Anthropocene hybridization events (Kumar et al., 2017; Kutschera et al., 2014). This raises the question whether conservationists can apply insights from these past hybridization events to inform current policy.
One could compare past introgression patterns with the current dynamics of the human‐mediated hybridization event, potentially allowing researchers to assess different outcomes with greater confidence. For example, according to one theoretical model of genome evolution during speciation, genomic islands of differentiation that house barrier loci are expected to increase in size as genomic regions are linked together. This process—known as genome hitchhiking—leads to an average genome‐wide reduction in introgression, ultimately culminating in complete reproductive isolation (Feder et al., 2012; Flaxman et al., 2013). Comparing the genomic landscape between pre‐ and post‐Anthropocene hybridization events might reveal this expansion of genomic islands over time, suggesting that reproductive isolation has been strengthened and genetic swamping is less likely to occur. It is also possible that some taxa are in merging‐and‐diverging cycles where periods of geographic isolation are punctuated by introgression events (Grant & Grant, 2008; McKay & Zink, 2015). During each allopatric phase, more genetic divergence builds up between the taxa, resulting in lower levels of introgression during the subsequent merging phase. Human‐mediated changes might have sped up the occurrence of a merging event, such as the interbreeding of polar bears and grizzly bears. Reconstructing the dynamics during these merging‐and‐diverging cycles might provide insights into the likelihood of genetic swamping or species collapse during the anthropogenic introgression event. However, the environmental and genetic context of pre‐Anthropocene hybridization events might not be comparable with the dynamics in current hybrid zones (Gompert et al., 2017). To my knowledge, no study has explicitly compared pre‐Anthropocene hybridization events with current human‐induced cases. This knowledge gap provides an exciting avenue for further research that could lead to better conservation measures.
8. RETICULATION AND CONSERVATION
The genetic legacy of past hybridization events is still detectable in present‐day genomes (Lombal et al., 2020; Ottenburghs, 2020; Taylor & Larson, 2019). These genetic patterns can be especially apparent in phylogenomic studies where phylogenetic analyses of different genomic regions point to distinct evolutionary histories—as one would expect based on the genic view of introgression (Edelman et al., 2019; Li et al., 2019; Ottenburghs, Kraus, et al., 2017; Ottenburghs, Megens, et al., 2017). For instance, phylogenomic analyses of the cat family (Felidae) revealed that the phylogenetic signal for the species tree was concentrated within genomic regions of low recombination, whereas regions of high recombination were heavily influenced by past introgression (Li et al., 2019). Apart from introgressive hybridization, this phylogenetic incongruence between different genomic regions—known as gene tree discordance—can also be the outcome of other evolutionary processes, such as incomplete lineage sorting or gene duplications (Degnan & Rosenberg, 2009; Maddison, 1997). Several methods have been developed to discriminate between these processes and to deal with high levels phylogenetic incongruence (Kapli et al., 2020; Ottenburghs, Kraus, et al., 2017; Ottenburghs, Megens, et al., 2017; Zhou et al., 2020). One promising approach is the application of phylogenetic networks to highlight the reticulated nature of evolution (Blair & Ané, 2019; Ottenburghs et al., 2016).
Conservation efforts are still mainly focused on a tree‐like pattern of evolution in which distinct evolutionary lineages warrant protection. This perspective is reflected in the Endangered Species Act (ESA) in the United States which lists and protects vulnerable species and subspecies of plants and animals (Ellstrand et al., 2010). At the moment, interspecific hybrids are generally denied protection under ESA (although some hybrid plant species might be considered). Similarly, the IUCN Red list does not consider hybrids (IUCN, 2020). Other organizations do take hybrids into account or provide guidelines on how to deal with hybridization (Jackiw et al., 2015). For example, the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) has included some hybrids in Appendices I and II (i.e., species threatened with extinction or species that will become threatened without controlling trade). These examples clearly indicate that there are still large discrepancies in conservation efforts regarding hybrids between different countries and organizations. Acknowledging the reticulated nature of the evolutionary process by reconstructing past hybridization events and adopting a phylogenetic network approach can help to design better guidelines for hybrids (vonHoldt et al., 2018).
Jackiw et al., (2015) proposed an elaborate framework to implement conservation efforts for hybrids, taking into account ethical and ecological considerations. We can incorporate insights from speciation genomics into this framework to deal with hybridization events in the Anthropocene (Figure 3). First, consider human‐mediated hybridization events (e.g., due to land‐use changes or the release of particular species). The release of individuals can be intentional as part of a genetic rescue program. If this program is closely monitored and there are no negative effects of hybridization, the hybrids are eligible for legal protection (Frankham, 2015). However, if the genetic rescue program leads to unforeseen issues, such as outbreeding depression or maladaptive introgression, and the hybrids threaten the endangered population, conservation efforts are needed (Frankham et al., 2011; Mills & Allendorf, 1996). This scenario also applies to the intentional release of game birds or fish that might negatively affect local populations through interbreeding (Randi, 2008). When the introduction of a particular species is unintentional—either due to individuals escaping from captivity or the spread of populations due to land‐use changes—the legal status of the hybrids depends on the native distribution of hybridizing species. If one parental species is non‐native, the hybrids cannot be legally protected and conservation measures should be implemented. For example, the introduction of North American Corbicula clams in the European river Rhine (Pfenninger et al., 2002). However, some studies documented adaptive introgression between a native and an exotic species (Oziolor et al., 2019; Valencia‐Montoya et al., 2020), indicating this possibility should be taken into account when setting the legal status of hybrids. When both parental species are native, the hybridization event can be treated as a natural phenomenon and the conservation status of the interbreeding species should be taken into account. If one or both species are threatened by genetic or demographic swamping, then legal protection of the resulting hybrids is not advisable and conservation efforts should be implemented. If the parental species are not threatened by hybridization, the situation needs to be assessed to determine whether hybridization could be beneficial (e.g., adaptive introgression or increasing genetic diversity). Here, insights from speciation genomics can be useful. However, be aware that this decision tree only provides a rough framework and each hybridization event—anthropogenic or natural—should be assessed individually (Allendorf et al., 2001).
FIGURE 3.
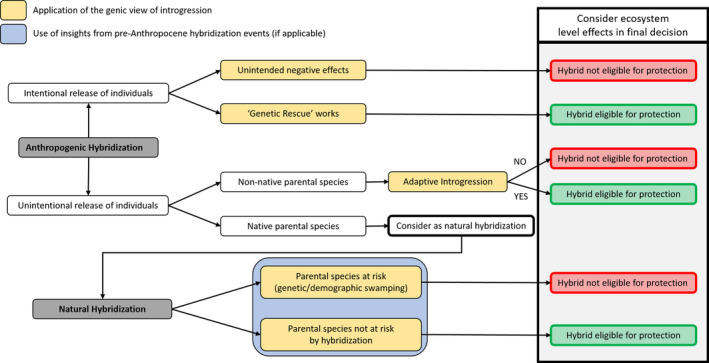
A decision tree to guide conservationists in determining the legal status of hybrids (based on Jackiw et al., 2015). Insights from speciation genomics (yellow) and past hybridization (blue) events are especially relevant in determining the risk for parental populations and the likelihood of adaptive introgression. In the final legal decision, it is important to also take into account potential ecosystem‐level effects of the hybrids. Please note that this decision tree only provides a rough framework and each hybridization event—anthropogenic or natural—should be assessed individually
An aspect that is currently missing from this framework is how anthropogenic hybridization affects ecosystem functioning. Most research on hybridization has focused on the interactions and genetic consequences between two or several taxa (Ottenburghs, 2019; Schwenk et al., 2008; Taylor & Larson, 2019). However, the production of hybrids not only affects the interbreeding taxa, it can also have far‐reaching consequences on an ecosystem level, altering food webs or nutrient cycles (Brennan et al., 2014). Some of these consequences are clearly negative, such as the spread of invasive pests (Corrêa et al., 2019) and the emergence of novel pathogens (Stukenbrock, 2016). But hybrids can also have a positive effect on other species in the ecosystem. For example, the spread of invasive Spartina hybrids provided extra habitat for the endangered California Ridgway's Rail (Rallus obsoletus) in the San Francisco Estuary (Ort & Thornton, 2016). Should the positive effect of these hybrids be taken into account in conservation efforts? The ecosystem perspective on anthropogenic hybridization adds another layer of complexity to the implementation of conservation measures.
9. CONCLUSION
As humans continue to change the environment and alter species distributions, more anthropogenic hybridization events will definitely occur. This will pose challenges for the conservation of endangered species, but also provide unique opportunities for evolutionary biologists. The interplay between conservation genomics and speciation genomics provides an exciting avenue for further research to gain important insights into the origin of species and how to protect them. In this review, I have illustrated this exchange of ideas by showing how insights from speciation genomics can guide the management of anthropogenic hybridization events. These insights range from practical issues (e.g., the development of diagnostic markers) to theoretical considerations, such as patterns of introgression, ancient hybridization events, and the reticulated nature of evolution.
From a conservation perspective, the unpredictability of (adaptive) introgression dynamics (Taylor & Larson, 2019) in combination with the strong link between anthropogenic hybridization and extinction risk (Todesco et al., 2016) suggests that the default course of action for human‐induced hybridization events should be the implementation of conservation measures to prevent hybridization. However, some studies documented the exchange of adaptive traits between native and non‐native species (Oziolor et al., 2019; Valencia‐Montoya et al., 2020), indicating that anthropogenic hybridization can result in adaptive introgression. The removal of introduced species and hybrids might have prevented the local species from adapting to the changing environment. These examples illustrate that each case of human‐induced hybridization should be judged separately (Allendorf et al., 2001). In addition, speciation genomic studies have shown that species can remain distinct in the face of high levels of introgression when species‐specific loci are not exchanged. Hence, high levels of introgression do not necessarily imply genetic swamping. It is not only important to quantify how much genetic material introgressed, but also what genomic regions have been exchanged. Finally, comparing current introgression dynamics with pre‐Anthropocene hybridization events might lead to novel insights on how to manage human‐mediated hybridization events, although this idea remains to be tested. Taking all these perspectives into account, in combination with the conservation status of the hybridizing taxa, thoughtful and evidence‐based conservation measures can be implemented.
CONFLICT OF INTEREST
None declared.
ACKNOWLEDGEMENTS
I would like to thank Frank Sterck, Pim van Hooft and two anonymous reviewers for their insightful comments on previous versions of this paper. And I thank Christophe Eizaguirre, Miguel Soares and Kristien Brans for reaching out and providing me the opportunity to contribute to this special issue on human‐induced evolution.
Ottenburghs J. The genic view of hybridization in the Anthropocene. Evolutionary Applications. 2021;14:2342–2360. 10.1111/eva.13223
DATA AVAILABILITY STATEMENT
There are not data associated with this manuscript, which is solely based on literature.
REFERENCES
- Alexander, D. , Novembre, J. , & Lange, K. (2009). Fast model‐based estimation of ancestry in unrelated individuals. Genome Research, 19(9), 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, T. J. , Vonlanthen, P. , & Seehausen, O. (2017). Does eutrophication‐driven evolution change aquatic ecosystems? Philosophical Transactions of the Royal Society B: Biological Sciences, 372(1712), 20160041. 10.1098/rstb.2016.0041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allendorf, F. W. , Leary, R. F. , Spruell, P. , & Wenburg, J. K. (2001). The problems with hybrids: Setting conservation guidelines. Trends in Ecology and Evolution, 16(11), 613–622. 10.1016/S0169-5347(01)02290-X [DOI] [Google Scholar]
- Alric, B. , Möst, M. , Domaizon, I. , Pignol, C. , Spaak, P. , & Perga, M.‐E. (2016). Local human pressures influence gene flow in a hybridizing Daphnia species complex. Journal of Evolutionary Biology, 29(4), 720–735. 10.1111/jeb.12820 [DOI] [PubMed] [Google Scholar]
- Anderson, D. , Toma, R. , Negishi, Y. , Okuda, K. , Ishiniwa, H. , Hinton, T. G. , Nanba, K. , Tamate, H. B. , & Kaneko, S. (2019). Mating of escaped domestic pigs with wild boar and possibility of their offspring migration after the Fukushima Daiichi Nuclear Power Plant accident. Scientific Reports, 9(1), 1–6. 10.1038/s41598-019-47982-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, E. (1948). Hybridization of the habitat. Evolution, 2, 1–9. [Google Scholar]
- Anderson, E. C. (2008). Bayesian inference of species hybrids using multilocus dominant genetic markers. Philosophical Transactions of the Royal Society B: Biological Sciences, 363(1505), 2841–2850. 10.1098/rstb.2008.0043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, T. M. , vonHoldt, B. M. , Candille, S. I. , Musiani, M. , Greco, C. , Stahler, D. R. , Smith, D. W. , Padhukasahasram, B. , Randi, E. , Leonard, J. A. , Bustamante, C. D. , Ostrander, E. A. , Tang, H. , Wayne, R. K. , & Barsh, G. S. (2009). Molecular and evolutionary history of melanism in North American gray wolves. Science, 323(5919), 1339–1343. 10.1126/science.1165448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, K. R. , Good, J. M. , Miller, M. R. , Luikart, G. , & Hohenlohe, P. A. (2016). Harnessing the power of RADseq for ecological and evolutionary genomics. Nature Reviews Genetics, 17(2), 81–92. 10.1038/nrg.2015.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, B. , Corbett‐Detig, R. B. , Hartl, D. , & Bomblies, K. (2013). RADseq underestimates diversity and introduces genealogical biases due to nonrandom haplotype sampling. Molecular Ecology, 22(11), 3179–3190. 10.1111/mec.12276 [DOI] [PubMed] [Google Scholar]
- Arnold, M. (1997). Natural hybridization and evolution. Oxford University Press. [Google Scholar]
- Arnold, M. , Ballerini, E. , & Brothers, A. (2012). Hybrid fitness, adaptation and evolutionary diversification: Lessons learned from Louisiana Irises. Heredity, 108(3), 159–166. 10.1038/hdy.2011.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, M. L. , & Kunte, K. (2017). Adaptive genetic exchange: A tangled history of admixture and evolutionary innovation. Trends in Ecology & Evolution, 32(8), 601–611. 10.1016/J.TREE.2017.05.007 [DOI] [PubMed] [Google Scholar]
- Arnold, M. , & Martin, N. (2010). Hybrid fitness across time and habitats. Trends in Ecology and Evolution, 25(9), 530–536. 10.1016/j.tree.2010.06.005 [DOI] [PubMed] [Google Scholar]
- Bangs, M. R. , Douglas, M. R. , Thompson, P. , & Douglas, M. E. (2017). Anthropogenic impacts facilitate native fish hybridization in the Bonneville Basin of western North America. Transactions of the American Fisheries Society, 146(1), 16–21. 10.1080/00028487.2016.1235611 [DOI] [Google Scholar]
- Bangs, M. R. , Oswald, K. J. , Greig, T. W. , Leitner, J. K. , Rankin, D. M. , & Quattro, J. M. (2018). Introgressive hybridization and species turnover in reservoirs: A case study involving endemic and invasive basses (Centrarchidae: Micropterus) in southeastern North America. Conservation Genetics, 19(1), 57–69. 10.1007/s10592-017-1018-7 [DOI] [Google Scholar]
- Battey, C. J. (2019). Evidence of linked selection on the z chromosome of hybridizing hummingbirds. Evolution, 74(4), 725–739. 10.1111/evo.13888 [DOI] [PubMed] [Google Scholar]
- Baveja, P. , Tang, Q. , Lee, J. G. H. , & Rheindt, F. E. (2019). Impact of genomic leakage on the conservation of the endangered Milky Stork. Biological Conservation, 229, 59–66. 10.1016/J.BIOCON.2018.11.009 [DOI] [Google Scholar]
- Bazykin, A. D. (1969). Hypothetical mechanism of speciation. Evolution, 23, 685–687. [DOI] [PubMed] [Google Scholar]
- Beheregaray, L. B. , Pfeiffer, L. V. , Attard, C. R. M. , Sandoval‐Castillo, J. , Domingos, F. M. C. B. , Faulks, L. K. , Gilligan, D. M. , & Unmack, P. J. (2017). Genome‐wide data delimits multiple climate‐determined species ranges in a widespread Australian fish, the golden perch (Macquaria ambigua). Molecular Phylogenetics and Evolution, 111, 65–75. 10.1016/j.ympev.2017.03.021 [DOI] [PubMed] [Google Scholar]
- Bell, R. C. , & Irian, C. G. (2019). Phenotypic and genetic divergence in reed frogs across a mosaic hybrid zone on São Tomé Island. Biological Journal of the Linnean Society, 128(3), 672–680. 10.1093/biolinnean/blz131 [DOI] [Google Scholar]
- Blair, C. , & Ané, C. (2019). Phylogenetic trees and networks can serve as powerful and complementary approaches for analysis of genomic data. Systematic Biology, 69(3), 593–601. 10.1093/sysbio/syz056 [DOI] [PubMed] [Google Scholar]
- Boecklen, W. J. , & Howard, D. J. (1997). Genetic analysis of hybrid zones: Numbers of markers and power of resolution. Ecology, 78(8), 2611–2616. [Google Scholar]
- Bourgeois, Y. X. , Bertrand, J. A. , Delahaie, B. , Holota, H. , Thébaud, C. , & Milá, B. (2020). Differential divergence in autosomes and sex chromosomes is associated with intra‐island diversification at a very small spatial scale in a songbird lineage. Molecular Ecology, 29(6), 1137–1153. 10.1111/mec.15396 [DOI] [PubMed] [Google Scholar]
- Bourke, P. M. , Voorrips, R. E. , Visser, R. G. F. , & Maliepaard, C. (2018). Tools for genetic studies in experimental populations of polyploids. Frontiers in Plant Science, 9, 513. 10.3389/fpls.2018.00513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan, A. C. , Woodward, G. , Seehausen, O. , Muñoz‐Fuentes, V. , Moritz, C. , Guelmami, A. , Abbott, R. J. , & Edelaar, P. (2014). Hybridization due to changing species distributions: Adding problems or solutions to conservation of biodiversity during global change? Evolutionary Ecology Research, 16(6), 475–491. [Google Scholar]
- Bruce, S. A. , Kutsumi, Y. , Van Maaren, C. , & Hare, M. P. (2020). Stocked‐fish introgression into wild brook trout populations depends on habitat. Transactions of the American Fisheries Society, 149(4), 427–442. 10.1002/tafs.10239 [DOI] [Google Scholar]
- Bruni, I. , De Mattia, F. , Fluch, S. , Ferrari, C. , Corazza, M. , Dinelli, E. , & Labra, M. (2016). Genetic introgression of hybrid Rhododendron x intermedium Tausch is habitat mediated: Evidences from south‐eastern Alps (Italy). Plant Biosystems, 150(3), 449–458. 10.1080/11263504.2014.986246 [DOI] [Google Scholar]
- Burri, R. (2017). Dissecting differentiation landscapes: A linked selection's perspective. Journal of Evolutionary Biology, 30(8), 1501–1505. 10.1111/jeb.13108 [DOI] [PubMed] [Google Scholar]
- Calabrese, G. M. , & Pfennig, K. S. (2020). Reinforcement and the proliferation of species. Journal of Heredity, 111(1), 138–146. 10.1093/jhered/esz073 [DOI] [PubMed] [Google Scholar]
- Campbell, C. R. , Poelstra, J. W. , & Yoder, A. D. (2018). What is Speciation Genomics? The roles of ecology, gene flow, and genomic architecture in the formation of species. Biological Journal of the Linnean Society, 124(4), 561–583. [Google Scholar]
- Canavan, K. , Paterson, I. D. , Lambertini, C. , & Hill, M. P. (2018). Expansive reed populations—Alien invasion or disturbed wetlands? AoB Plants, 10(2), ply014. 10.1093/aobpla/ply014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carantón‐Ayala, D. , Avendaño, J. E. , & Cadena, C. D. (2018). Hybridization in brushfinches (Atlapetes, Emberizidae) from the southeast Andes of Colombia: A consequence of habitat disturbance? Journal of Ornithology, 159(3), 713–722. 10.1007/s10336-018-1544-1 [DOI] [Google Scholar]
- Carney, S. E. , Gardner, K. A. , & Rieseberg, L. H. (2000). Evolutionary changes over the fifty‐year history of a hybrid population of sunflowers (Helianthus). Evolution, 54(2), 462–474. 10.1111/j.0014-3820.2000.tb00049.x [DOI] [PubMed] [Google Scholar]
- Chafin, T. K. , Douglas, M. R. , Martin, B. T. , & Douglas, M. E. (2019). Hybridization drives genetic erosion in sympatric desert fishes of western North America. Heredity, 123(6), 759–773. 10.1038/s41437-019-0259-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, W. Y. , Hoffmann, A. A. , & Oppen, M. J. H. (2019). Hybridization as a conservation management tool. Conservation Letters, 12(5), e12652. 10.1111/conl.12652 [DOI] [Google Scholar]
- Chunco, A. J. (2014). Hybridization in a warmer world. Ecology and Evolution, 4(10), 2019–2031. 10.1002/ece3.1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, L. V. , Lipka, A. E. , & Sacks, E. J. (2019). polyRAD: Genotype calling with uncertainty from sequencing data in polyploids and diploids. G3: Genes, Genomes, Genetics, 9(3), 663–673. 10.1534/g3.118.200913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppi, A. , Baker, A. J. M. , Bettarini, I. , Colzi, I. , Echevarria, G. , Pazzagli, L. , Gonnelli, C. , & Selvi, F. (2020). Population Genetics of Odontarrhena (Brassicaceae) from Albania: The effects of anthropic habitat disturbance, soil, and altitude on a Ni‐Hyperaccumulator plant group from a major serpentine hotspot. Plants, 9(12), 1686. 10.3390/plants9121686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrêa, A. S. , Cordeiro, E. M. , & Omoto, C. (2019). Agricultural insect hybridization and implications for pest management. Pest Management Science, 75(11), 2857–2864. 10.1002/ps.5495 [DOI] [PubMed] [Google Scholar]
- Coughlan, J. M. , & Matute, D. R. (2020). The importance of intrinsic postzygotic barriers throughout the speciation process. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 375(1806), 20190533. 10.1098/rstb.2019.0533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, J. A. , & Orr, H. A. (1989). Patterns of speciation in Drosophila. Evolution, 43(2), 362–381. 10.1111/j.1558-5646.1989.tb04233.x [DOI] [PubMed] [Google Scholar]
- Crispo, E. , Moore, J. S. , Lee‐Yaw, J. A. , Gray, S. M. , & Haller, B. C. (2011). Broken barriers: Human‐induced changes to gene flow and introgression in animals: An examination of the ways in which humans increase genetic exchange among populations and species and the consequences for biodiversity. BioEssays, 33(7), 508–518. 10.1002/bies.201000154 [DOI] [PubMed] [Google Scholar]
- Crutzen, P. J. (2002). Geology of mankind. Nature, 415(6867), 23. 10.1038/415023a [DOI] [PubMed] [Google Scholar]
- Degnan, J. H. , & Rosenberg, N. A. (2009). Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends in Ecology & Evolution, 24(6), 332–340. 10.1016/J.TREE.2009.01.009 [DOI] [PubMed] [Google Scholar]
- Derry, A. M. , Fraser, D. J. , Brady, S. P. , Astorg, L. , Lawrence, E. R. , Martin, G. K. , Matte, J. M. , Negrín Dastis, J. O. , Paccard, A. , Barrett, R. D. H. , Chapman, L. J. , Lane, J. E. , Ballas, C. G. , Close, M. , & Crispo, E. (2019). Conservation through the lens of (mal)adaptation: Concepts and meta‐analysis. Evolutionary Applications, 12(7), 1287–1304. 10.1111/eva.12791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey, S. , Lavanchy, G. , Thiebaud, J. , & Dufresnes, C. (2019). Herps without borders: A new newt case and a review of transalpine alien introductions in western Europe. Amphibia Reptilia, 40(1), 13–27. 10.1163/15685381-20181028 [DOI] [Google Scholar]
- Dufresnes, C. , Leuenberger, J. , Amrhein, V. , Bühler, C. , Thiébaud, J. , Bohnenstengel, T. , & Dubey, S. (2018). Invasion genetics of marsh frogs (Pelophylax ridibundus sensu lato) in Switzerland. Biological Journal of the Linnean Society, 123(2), 402–410. 10.1093/biolinnean/blx140 [DOI] [Google Scholar]
- Edelman, N. B. , Frandsen, P. B. , Miyagi, M. , Clavijo, B. , Davey, J. , Dikow, R. B. , García‐Accinelli, G. , Van Belleghem, S. M. , Patterson, N. , Neafsey, D. E. , Challis, R. , Kumar, S. , Moreira, G. R. P. , Salazar, C. , Chouteau, M. , Counterman, B. A. , Papa, R. , Blaxter, M. , Reed, R. D. , … Mallet, J. (2019). Genomic architecture and introgression shape a butterfly radiation. Science, 366(6465), 594–599. 10.1126/science.aaw2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmands, S. (1999). Heterosis and outbreeding depression in interpopulation crosses spanning a wide range of divergence. Evolution, 53(6), 1757–1768. 10.1111/j.1558-5646.1999.tb04560.x [DOI] [PubMed] [Google Scholar]
- Ellstrand, N. C. , Biggs, D. , Kaus, A. , Lubinsky, P. , McDade, L. A. , Preston, K. , Prince, L. M. , Regan, H. M. , Rorive, V. , Ryder, O. A. , & Schierenbeck, K. A. (2010). Got hybridization? A multidisciplinary approach for informing science policy. BioScience, 60(5), 384–388. 10.1525/bio.2010.60.5.8 [DOI] [Google Scholar]
- Escalante, M. A. , Perrier, C. , García‐De León, F. J. , Ruiz‐Luna, A. , Ortega‐Abboud, E. , & Manel, S. (2020). Genotyping‐by‐sequencing reveals the effects of riverscape, climate and interspecific introgression on the genetic diversity and local adaptation of the endangered Mexican golden trout (Oncorhynchus chrysogaster). Conservation Genetics, 21(5), 907–926. 10.1007/s10592-020-01297-z [DOI] [Google Scholar]
- Fadakar, D. , Malekian, M. , Hemami, M. R. , Lerp, H. , Rezaei, H. R. , & Bärmann, E. V. (2020). Repeated hybridization of two closely related gazelle species (Gazella bennettii and Gazella subgutturosa) in central Iran. Ecology and Evolution, 10(20), 11372–11386. 10.1002/ece3.6774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulks, L. , & Östman, Ö. (2016). Genetic Diversity and hybridisation between native and introduced Salmonidae fishes in a Swedish Alpine Lake. PLoS One, 11(3), e0152732. 10.1371/journal.pone.0152732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder, J. L. , Egan, S. P. , & Nosil, P. (2012). The genomics of speciation‐with‐gene‐flow. Trends in Genetics, 28(7), 342–350. [DOI] [PubMed] [Google Scholar]
- Feulner, P. G. D. , & Seehausen, O. (2019). Genomic insights into the vulnerability of sympatric whitefish species flocks. Molecular Ecology, 28(3), 615–629. 10.1111/mec.14977 [DOI] [PubMed] [Google Scholar]
- Ficetola, G. F. , Lunghi, E. , Cimmaruta, R. , & Manenti, R. (2019). Transgressive niche across a salamander hybrid zone revealed by microhabitat analyses. Journal of Biogeography, 46(7), jbi.13621. 10.1111/jbi.13621 [DOI] [Google Scholar]
- Fisher, H. S. , Wong, B. B. , & Rosenthal, G. G. (2006). Alteration of the chemical environment disrupts communication in a freshwater fish. Proceedings of the Royal Society B: Biological Sciences, 273(1591), 1187–1193. 10.1098/rspb.2005.3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaxman, S. M. , Feder, J. L. , & Nosil, P. (2013). Genetic hitchhiking and the dynamic buildup of genomic divergence during speciation with gene flow. Evolution, 67(9), 2577–2591. 10.1111/evo.12055 [DOI] [PubMed] [Google Scholar]
- Forcina, G. , Guerrini, M. , & Barbanera, F. (2020). Non‐native and hybrid in a changing environment: Conservation perspectives for the last Italian red‐legged partridge (Alectoris rufa) population with long natural history. Zoology, 138, 125740. 10.1016/j.zool.2019.125740 [DOI] [PubMed] [Google Scholar]
- Forcina, G. , Guerrini, M. , Khaliq, I. , Khan, A. A. , & Barbanera, F. (2018). Human‐modified biogeographic patterns and conservation in game birds: The dilemma of the black francolin (Francolinus francolinus, Phasianidae) in Pakistan. PLoS One, 13(10), e0205059. 10.1371/journal.pone.0205059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham, R. (2015). Genetic rescue of small inbred populations: Meta‐analysis reveals large and consistent benefits of gene flow. Molecular Ecology, 24(11), 2610–2618. 10.1111/mec.13139 [DOI] [PubMed] [Google Scholar]
- Frankham, R. , Ballou, J. D. , Eldridge, M. D. B. , Lacy, R. C. , Ralls, K. , Dudash, M. R. , & Fenster, C. B. (2011). Predicting the probability of outbreeding depression. Conservation Biology, 25(3), 465–475. 10.1111/j.1523-1739.2011.01662.x [DOI] [PubMed] [Google Scholar]
- Fraser, G. S. , DeHaan, P. W. , Smith, C. T. , Von Bargen, J. F. , Cooper, M. R. , & Desgrosseillier, T. J. (2020). Overlap of spatial and temporal spawning distributions of spring and summer Chinook Salmon results in hybridization in the upper Columbia River. Transactions of the American Fisheries Society, 149(5), 517–531. 10.1002/tafs.10258 [DOI] [Google Scholar]
- Funk, W. C. , Forester, B. R. , Converse, S. J. , Darst, C. , & Morey, S. (2019). Improving conservation policy with genomics: A guide to integrating adaptive potential into U.S. Endangered Species Act decisions for conservation practitioners and geneticists. Conservation Genetics, 20(1), 115–134. 10.1007/s10592-018-1096-1 [DOI] [Google Scholar]
- Galaverni, M. , Caniglia, R. , Pagani, L. , Fabbri, E. , Boattini, A. , & Randi, E. (2017). Disentangling timing of admixture, patterns of introgression, and phenotypic indicators in a hybridizing wolf population. Molecular Biology and Evolution, 34(9), 2324–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmendia, A. , Merle, H. , Ruiz, P. , & Ferriol, M. (2018). Distribution and ecological segregation on regional and microgeographic scales of the diploid Centaurea aspera L., the tetraploid C. seridis L., and their triploid hybrids (Compositae). PeerJ, 2018(7), e5209. 10.7717/peerj.5209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garroway, C. J. , Bowman, J. , Cascaden, T. J. , Holloway, G. L. , Mahan, C. G. , Malcolm, J. R. , Steele, M. A. , Turner, G. , & Wilson, P. J. (2010). Climate change induced hybridization in flying squirrels. Global Change Biology, 16(1), 113–121. 10.1111/j.1365-2486.2009.01948.x [DOI] [Google Scholar]
- Gibson, I. , Welsh, A. B. , Welsh, S. A. , & Cincotta, D. A. (2019). Genetic swamping and possible species collapse: Tracking introgression between the native Candy Darter and introduced Variegate Darter. Conservation Genetics, 20(2), 287–298. 10.1007/s10592-018-1131-2 [DOI] [Google Scholar]
- Glasheen, P. M. , Burks, R. L. , Campos, S. R. , & Hayes, K. A. (2020). First evidence of introgressive hybridization of apple snails (Pomacea spp.) in their native range. Journal of Molluscan Studies, 86(2), 96–103. 10.1093/mollus/eyz035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompert, Z. , Mandeville, E. G. , & Buerkle, C. A. (2017). Analysis of population genomic data from hybrid zones. Annual Review of Ecology, Evolution, and Systematics, 48, 207–229. 10.1146/annurev-ecolsys-110316-022652 [DOI] [Google Scholar]
- Gong, S. , Vamberger, M. , Auer, M. , Praschag, P. , & Fritz, U. (2018). Millennium‐old farm breeding of Chinese softshell turtles (Pelodiscus spp.) results in massive erosion of biodiversity. The Science of Nature, 105(5–6), 1–10. 10.1007/s00114-018-1558-9 [DOI] [PubMed] [Google Scholar]
- Grabenstein, K. C. , & Taylor, S. A. (2018). Breaking barriers: Causes, consequences, and experimental utility of human‐mediated hybridization. Trends in Ecology and Evolution, 33(3), 198–212. 10.1016/j.tree.2017.12.008 [DOI] [PubMed] [Google Scholar]
- Graham, C. F. , Eberts, R. L. , Goncin, U. , & Somers, C. M. (2020). Spontaneous hybridization and introgression between walleye (Sander vitreus) and sauger (Sander canadensis) in two large reservoirs: Insights from genotyping by sequencing. Evolutionary Applications. 10.1111/eva.13174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, B. R. , & Grant, P. R. (2008). Fission and fusion of Darwin’s finches populations. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 363(1505), 2821–2829. 10.1098/rstb.2008.0051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobler, P. , van Wyk, A. M. , Dalton, D. L. , van Vuuren, B. J. , & Kotzé, A. (2018). Assessing introgressive hybridization between blue wildebeest (Connochaetes taurinus) and black wildebeest (Connochaetes gnou) from South Africa. Conservation Genetics, 19(4), 981–993. 10.1007/s10592-018-1071-x [DOI] [Google Scholar]
- Guivier, E. , Gilles, A. , Pech, N. , Duflot, N. , Tissot, L. , & Chappaz, R. (2019). Canals as ecological corridors and hybridization zones for two cyprinid species. Hydrobiologia, 830(1), 1–16. 10.1007/s10750-018-3843-1 [DOI] [Google Scholar]
- Harrisson, K. A. , Pavlova, A. , Gonçalves da Silva, A. , Rose, R. , Bull, J. K. , Lancaster, M. L. , Murray, N. , Quin, B. , Menkhorst, P. , Magrath, M. J. L. , & Sunnucks, P. (2016). Scope for genetic rescue of an endangered subspecies though re‐establishing natural gene flow with another subspecies. Molecular Ecology, 25(6), 1242–1258. 10.1111/mec.13547 [DOI] [PubMed] [Google Scholar]
- Heber, S. , Varsani, A. , Kuhn, S. , Girg, A. , Kempenaers, B. , & Briskie, J. (2013). The genetic rescue of two bottlenecked South Island robin populations using translocations of inbred donors. Proceedings of the Royal Society B: Biological Sciences, 280(1752), 20122228. 10.1098/rspb.2012.2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick, P. W. (2013). Adaptive introgression in animals: Examples and comparison to new mutation and standing variation as sources of adaptive variation. Molecular Ecology, 22(18), 4606–4618. 10.1111/mec.12415 [DOI] [PubMed] [Google Scholar]
- Hibbins, M. , & Hahn, M. (2021). Phylogenomic approaches to detecting and characterizing introgression. EcoEvoRxiv. 10.32942/OSF.IO/UAHD8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, P. J. (2018). Changes in native bull trout and non‐native brook trout distributions in the upper Powder River basin after 20 years, relationships to water temperature and implications of climate change. Ecology of Freshwater Fish, 27(3), 710–719. 10.1111/eff.12386 [DOI] [Google Scholar]
- Huuskonen, H. , Shikano, T. , Mehtätalo, L. , Kettunen, J. , Eronen, R. , Toiviainen, A. , & Kekäläinen, J. (2017). Anthropogenic environmental changes induce introgression in sympatric whitefish ecotypes. Biological Journal of the Linnean Society, 121(3), 613–626. 10.1093/biolinnean/blx010 [DOI] [Google Scholar]
- IUCN . (2020). IUCN Red List of Threatened Species. Retrieved from https://www.iucnredlist.org/resources/tax‐sources [Google Scholar]
- Jackiw, R. N. , Mandil, G. , & Hager, H. A. (2015). A framework to guide the conservation of species hybrids based on ethical and ecological considerations. Conservation Biology, 29(4), 1040–1051. 10.1111/cobi.12526 [DOI] [PubMed] [Google Scholar]
- Jones, M. R. , Mills, L. S. , Alves, P. C. , Callahan, C. M. , Alves, J. M. , Lafferty, D. J. R. , Jiggins, F. M. , Jensen, J. D. , Melo‐Ferreira, J. , & Good, J. M. (2018). Adaptive introgression underlies polymorphic seasonal camouflage in snowshoe hares. Science, 360(6395), 1355–1358. 10.1126/science.aar5273 [DOI] [PubMed] [Google Scholar]
- Kapli, P. , Yang, Z. , & Telford, M. J. (2020). Phylogenetic tree building in the genomic age. Nature Reviews Genetics, 21, 428–444. 10.1038/s41576-020-0233-0 [DOI] [PubMed] [Google Scholar]
- Kelly, B. P. , Whiteley, A. , & Tallmon, D. (2010). The Arctic melting pot. Nature, 468(7326), 891. 10.1038/468891a [DOI] [PubMed] [Google Scholar]
- Key, K. H. L. (1968). The concept of stasipatric speciation. Systematic Zoology, 17, 14–22. [Google Scholar]
- Kim, M. , Cui, M.‐L. , Cubas, P. , Gillies, A. , Lee, K. , Chapman, M. A. , Abbott, R. J. , & Coen, E. (2008). Regulatory genes control a key morphological and ecological trait transferred between species. Science, 322(5904), 1116–1119. 10.1126/science.1164371 [DOI] [PubMed] [Google Scholar]
- Kumar, V. , Lammers, F. , Bidon, T. , Pfenninger, M. , Kolter, L. , Nilsson, M. A. , & Janke, A. (2017). The evolutionary history of bears is characterized by gene flow across species. Scientific Reports, 7(1), 46487. 10.1038/srep46487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschera, V. E. , Bidon, T. , Hailer, F. , Rodi, J. L. , Fain, S. R. , & Janke, A. (2014). Bears in a forest of gene trees: Phylogenetic inference is complicated by incomplete lineage sorting and gene flow. Molecular Biology and Evolution, 31(8), 2004–2017. 10.1093/molbev/msu186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhaney, S. , Han, F. , Webster, M. T. , Andersson, L. , Grant, B. R. , & Grant, P. R. (2018). Rapid hybrid speciation in Darwin’s finches. Science, 359(6372), 224–228. 10.1126/science.aao4593 [DOI] [PubMed] [Google Scholar]
- Larson, E. L. , Tinghitella, R. M. , & Taylor, S. A. (2019). Insect hybridization and climate change. Frontiers in Ecology and Evolution, 7, 348. 10.3389/fevo.2019.00348 [DOI] [Google Scholar]
- Lau, C. L. F. , & Jacobs, D. K. (2017). Introgression between ecologically distinct species following increased salinity in the Colorado Delta‐ Worldwide implications for impacted estuary diversity. PeerJ, 2017(12), e4056. 10.7717/peerj.4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavretsky, P. , Janzen, T. , & McCracken, K. G. (2019). Identifying hybrids & the genomics of hybridization: Mallards & American black ducks of Eastern North America. Ecology and Evolution, 9(6), 3470–3490. 10.1002/ece3.4981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavretsky, P. , McInerney, N. R. , Mohl, J. E. , Brown, J. I. , James, H. F. , McCracken, K. G. , & Fleischer, R. C. (2020). Assessing changes in genomic divergence following a century of human mediated secondary contact among wild and captive‐bred ducks. Molecular Ecology, 29(3), 578–595. 10.1111/mec.15343 [DOI] [PubMed] [Google Scholar]
- Lemopoulos, A. , Prokkola, J. M. , Uusi‐Heikkilä, S. , Vasemägi, A. , Huusko, A. , Hyvärinen, P. , Koljonen, M. L. , Koskiniemi, J. , & Vainikka, A. (2019). Comparing RADseq and microsatellites for estimating genetic diversity and relatedness — Implications for brown trout conservation. Ecology and Evolution, 9(4), 2106–2120. 10.1002/ece3.4905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, S. L. , & Maslin, M. A. (2015). Defining the Anthropocene. Nature, 519(7542), 171–180. 10.1038/nature14258 [DOI] [PubMed] [Google Scholar]
- Li, G. , Figueiró, H. V. , Eizirik, E. , & Murphy, W. J. (2019). Recombination‐aware phylogenomics reveals the structured genomic landscape of hybridizing cat species. Molecular Biology and Evolution, 36(10), 2111–2126. 10.1093/molbev/msz139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombal, A. J. , O'dwyer, J. E. , Friesen, V. , Woehler, E. J. , & Burridge, C. P. (2020). Identifying mechanisms of genetic differentiation among populations in vagile species: Historical factors dominate genetic differentiation in seabirds. Biological Reviews, 95(3), 625–651. 10.1111/brv.12580 [DOI] [PubMed] [Google Scholar]
- Lowry, D. B. , Hoban, S. , Kelley, J. L. , Lotterhos, K. E. , Reed, L. K. , Antolin, M. F. , & Storfer, A. (2016). Breaking RAD: An evaluation of the utility of restriction site associated DNA sequencing for genome scans of adaptation. Molecular Ecology Resources, 17(2), 142–152. 10.1111/1755-0998.12596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison, W. P. (1997). Gene trees in species trees. Systematic Biology, 46(3), 523–536. 10.1093/sysbio/46.3.523 [DOI] [Google Scholar]
- Mallet, J. (2007). Hybrid speciation. Nature, 446(7133), 279–283. 10.1038/nature05706 [DOI] [PubMed] [Google Scholar]
- Malukiewicz, J. (2019). A review of experimental, natural, and anthropogenic hybridization in Callithrix marmosets. International Journal of Primatology, 40(1), 72–98. 10.1007/s10764-018-0068-0 [DOI] [Google Scholar]
- Mank, J. E. , Carlson, J. E. , & Brittingham, M. C. (2004). A century of hybridization: Decreasing genetic distance between American black ducks and mallards. Conservation Genetics, 5(3), 395–403. 10.1023/B:COGE.0000031139.55389.b1 [DOI] [Google Scholar]
- Marques, D. A. , Meier, J. I. , & Seehausen, O. (2019). A combinatorial view on speciation and adaptive radiation. Trends in Ecology & Evolution, 34(6), 531–544.Retrieved from https://www.sciencedirect.com/science/article/pii/S0169534719300552 [DOI] [PubMed] [Google Scholar]
- Mayr, E. (1963). Animal species and evolution. Belknap Press of Harvard University Press. [Google Scholar]
- McFarlane, S. E. , Hunter, D. C. , Senn, H. V. , Smith, S. L. , Holland, R. , Huisman, J. , & Pemberton, J. M. (2020). Increased genetic marker density reveals high levels of admixture between red deer and introduced Japanese sika in Kintyre. Scotland. Evolutionary Applications, 13(2), 432–441. 10.1111/eva.12880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane, S. E. , & Pemberton, J. M. (2019). Detecting the true extent of introgression during anthropogenic hybridization. Trends in Ecology & Evolution, 34(4), 315–326. 10.1016/J.TREE.2018.12.013 [DOI] [PubMed] [Google Scholar]
- McKay, B. D. , & Zink, R. M. (2015). Sisyphean evolution in Darwin’s finches. Biological Reviews, 90(3), 689–698. 10.1111/brv.12127 [DOI] [PubMed] [Google Scholar]
- Miller, J. , Poissant, J. , Hogg, J. , & Coltman, D. (2012). Genomic consequences of genetic rescue in an insular population of bighorn sheep (Ovis canadensis). Molecular Ecology, 21(7), 1583–1596. 10.1111/j.1365-294X.2011.05427.x [DOI] [PubMed] [Google Scholar]
- Miller, S. M. , Moeller, C.‐H. , Harper, C. K. , & Bloomer, P. (2020). Anthropogenic movement results in hybridisation in impala in southern Africa. Conservation Genetics, 21, 653–663. 10.1007/s10592-020-01276-4 [DOI] [Google Scholar]
- Millette, K. L. , Gonzalez, A. , & Cristescu, M. E. (2020). Breaking ecological barriers: Anthropogenic disturbance leads to habitat transitions, hybridization, and high genetic diversity. Science of the Total Environment, 740, 140046. 10.1016/j.scitotenv.2020.140046 [DOI] [PubMed] [Google Scholar]
- Mills, L. S. , & Allendorf, F. W. (1996). The one‐migrant‐per‐generation rule in conservation and management. Conservation Biology, 10(6), 1509–1518. 10.1046/j.1523-1739.1996.10061509.x [DOI] [Google Scholar]
- Milot, E. , Béchet, A. , & Maris, V. (2020). The dimensions of evolutionary potential in biological conservation. Evolutionary Applications, 13(6), 1363–1379. 10.1111/eva.12995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moest, M. , Van Belleghem, S. M. , James, J. E. , Salazar, C. , Martin, S. H. , Barker, S. L. , Moreira, G. R. P. , Mérot, C. , Joron, M. , Nadeau, N. J. , Steiner, F. M. , & Jiggins, C. D. (2020). Selective sweeps on novel and introgressed variation shape mimicry loci in a butterfly adaptive radiation. PLOS Biology, 18(2), e3000597. 10.1371/journal.pbio.3000597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais, P. , & Reichard, M. (2018). Cryptic invasions: A review. Science of the Total Environment, 613, 1438–1448. 10.1016/j.scitotenv.2017.06.133 [DOI] [PubMed] [Google Scholar]
- Morii, K. , Nakano, M. , & Takakura, K. I. (2018). Does simultaneous and sympatric reproduction between two native spined loaches lead to reproductive interference and local extinction? Environmental Biology of Fishes, 101(9), 1407–1416. 10.1007/s10641-018-0787-2 [DOI] [Google Scholar]
- Moulton, L. L. , Vallender, R. , Artuso, C. , & Koper, N. (2017). The final frontier: Early‐stage genetic introgression and hybrid habitat use in the northwestern extent of the Golden‐winged Warbler breeding range. Conservation Genetics, 18(6), 1481–1487. 10.1007/s10592-017-0989-8 [DOI] [Google Scholar]
- Muhlfeld, C. C. , Kalinowski, S. T. , McMahon, T. E. , Taper, M. L. , Painter, S. , Leary, R. F. , & Allendorf, F. W. (2009). Hybridization rapidly reduces fitness of a native trout in the wild. Biology Letters, 5(3), 328–331. 10.1098/rsbl.2009.0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlfeld, C. C. , Kovach, R. P. , Al‐Chokhachy, R. , Amish, S. J. , Kershner, J. L. , Leary, R. F. , Lowe, W. H. , Luikart, G. , Matson, P. , Schmetterling, D. A. , Shepard, B. B. , Westley, P. A. H. , Whited, D. , Whiteley, A. , & Allendorf, F. W. (2017). Legacy introductions and climatic variation explain spatiotemporal patterns of invasive hybridization in a native trout. Global Change Biology, 23(11), 4663–4674. 10.1111/gcb.13681 [DOI] [PubMed] [Google Scholar]
- Nevado, B. , Harris, S. A. , Beaumont, M. A. , & Hiscock, S. J. (2020). Rapid homoploid hybrid speciation in British gardens: The origin of Oxford ragwort (Senecio squalidus). Molecular Ecology, 29(21), 4221–4233. 10.1111/mec.15630 [DOI] [PubMed] [Google Scholar]
- O’Donnell, R. P. , Drost, C. A. , & Mock, K. E. (2017). Cryptic invasion of Northern Leopard Frogs (Rana pipiens) across phylogeographic boundaries and a dilemma for conservation of a declining amphibian. Biological Invasions, 19(3), 1039–1052. 10.1007/s10530-016-1320-1 [DOI] [Google Scholar]
- Ort, B. S. , & Thornton, W. J. (2016). Changes in the population genetics of an invasive Spartina after 10 years of management. Biological Invasions, 18(8), 2267–2281. 10.1007/s10530-016-1177-3 [DOI] [Google Scholar]
- Ortego, J. , Gugger, P. F. , & Sork, V. L. (2017). Impacts of human‐induced environmental disturbances on hybridization between two ecologically differentiated Californian oak species. New Phytologist, 213(2), 942–955. 10.1111/nph.14182 [DOI] [PubMed] [Google Scholar]
- Oswald, J. A. , Harvey, M. G. , Remsen, R. C. , Foxworth, D. U. , Dittmann, D. L. , Cardiff, S. W. , & Brumfield, R. T. (2019). Evolutionary dynamics of hybridization and introgression following the recent colonization of Glossy Ibis (Aves: Plegadis falcinellus) into the New World. Molecular Ecology, 28(7), 1675–1691. 10.1111/MEC.15008 [DOI] [PubMed] [Google Scholar]
- Ottenburghs, J. (2018). Exploring the hybrid speciation continuum in birds. Ecology and Evolution, 8(24), 13027–13034. 10.1002/ece3.4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenburghs, J. (2019). Multispecies hybridization in birds. Avian Research, 10, 20. 10.1186/s40657-019-0159-4 [DOI] [Google Scholar]
- Ottenburghs, J. (2020). Ghost introgression: Spooky Gene flow in the distant past. BioEssays, 42(6), 2000012. 10.1002/bies.202000012 [DOI] [PubMed] [Google Scholar]
- Ottenburghs, J. , Honka, J. , Müskens, G. J. D. M. , & Ellegren, H. (2020). Recent introgression between Taiga Bean Goose and Tundra Bean Goose results in a largely homogeneous landscape of genetic differentiation. Heredity, 125(1–2), 73–84. 10.1038/s41437-020-0322-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenburghs, J. , Kraus, R. , van Hooft, P. , van Wieren, S. , Ydenberg, R. , & Prins, H. (2017). Avian introgression in the genomic era. Avian Research, 8(1), 30. [Google Scholar]
- Ottenburghs, J. , Megens, H.‐J. , Kraus, R. , Van Hooft, P. , Van Wieren, S. , Crooijmans, R. , Ydenberg, R. , Groenen, M. , & Prins, H. (2017). A history of hybrids? Genomic patterns of introgression in the True Geese. BMC Evolutionary Biology, 17(1), 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenburghs, J. , van Hooft, P. , van Wieren, S. , Ydenberg, R. , & Prins, H. (2016). Birds in a bush: Toward an avian phylogenetic network. The Auk, 133(4), 577–582. [Google Scholar]
- Oziolor, E. M. , Reid, N. M. , Yair, S. , Lee, K. M. , Guberman VerPloeg, S. , Bruns, P. C. , Shaw, J. R. , Whitehead, A. , & Matson, C. W. (2019). Adaptive introgression enables evolutionary rescue from extreme environmental pollution. Science, 364(6439), 455–457. 10.1126/science.aav4155 [DOI] [PubMed] [Google Scholar]
- Parmesan, C. (2006). Ecological and evolutionary responses to recent climate change. Annual Review of Ecology, Evolution, and Systematics, 37(1), 637–669. 10.1146/annurev.ecolsys.37.091305.110100 [DOI] [Google Scholar]
- Payseur, B. (2010). Using differential introgression in hybrid zones to identify genomic regions involved in speciation. Molecular Ecology Resources, 10(5), 806–820. 10.1111/j.1755-0998.2010.02883.x [DOI] [PubMed] [Google Scholar]
- Peng, Y. , Dong, Y. , Xu, H. , Xi, X. , Niklas, K. J. , & Sun, S. (2018). Domesticated honeybees facilitate interspecific hybridization between two Taraxacum congeners. Journal of Ecology, 106(3), 1204–1216. 10.1111/1365-2745.12909 [DOI] [Google Scholar]
- Peona, V. , Weissensteiner, M. , & Suh, A. (2018). How complete are ‘complete’ genome assemblies?‐An avian perspective. Molecular Ecology Resources, 18(6), 1188–1195. [DOI] [PubMed] [Google Scholar]
- Pfenninger, M. , Reinhardt, F. , & Streit, B. (2002). Evidence for cryptic hybridization between different evolutionary lineages of the invasive clam genus Corbicula (Veneroida, Bivalvia). Journal of Evolutionary Biology, 15(5), 818–829. 10.1046/j.1420-9101.2002.00440.x [DOI] [Google Scholar]
- Pickup, M. , Field, D. L. , Rowell, D. M. , & Young, A. G. (2013). Source population characteristics affect heterosis following genetic rescue of fragmented plant populations. Proceedings of the Royal Society B: Biological Sciences, 280(1750), 20122058. 10.1098/rspb.2012.2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimm, S. L. , Dollar, L. , & Bass, O. L. (2006). The genetic rescue of the Florida panther. Animal Conservation, 9(2), 115–122. 10.1111/j.1469-1795.2005.00010.x [DOI] [Google Scholar]
- Plomion, C. , Aury, J.‐M. , Amselem, J. , Leroy, T. , Murat, F. , Duplessis, S. , Faye, S. , Francillonne, N. , Labadie, K. , Le Provost, G. , Lesur, I. , Bartholomé, J. , Faivre‐Rampant, P. , Kohler, A. , Leplé, J.‐C. , Chantret, N. , Chen, J. , Diévart, A. , Alaeitabar, T. , … Salse, J. (2018). Oak genome reveals facets of long lifespan. Nature Plants, 4(7), 440–452. 10.1038/s41477-018-0172-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa‐Báez, Á. D. , Catullo, R. , Lee, S. F. , Yeap, H. L. , Mourant, R. G. , Frommer, M. , Sved, J. A. , Cameron, E. C. , Edwards, O. R. , Taylor, P. W. , & Oakeshott, J. G. (2020). Genome‐wide patterns of differentiation over space and time in the Queensland fruit fly. Scientific Reports, 10(1), 1–13. 10.1038/s41598-020-67397-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentis, P. J. , White, E. M. , Radford, I. J. , Lowe, A. J. , & Clarke, A. R. (2007). Can hybridization cause local extinction: A case for demographic swamping of the Australian native Senecio pinnatifolius by the invasive Senecio madagascariensis? New Phytologist, 176(4), 902–912. 10.1111/j.1469-8137.2007.02217.x [DOI] [PubMed] [Google Scholar]
- Presgraves, D. C. (2002). Patterns of postzygotic isolation in Lepidoptera. Evolution, 56(6), 1168–1183. 10.1111/j.0014-3820.2002.tb01430.x [DOI] [PubMed] [Google Scholar]
- Pritchard, J. K. , Stephens, M. , & Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics, 155(2), 945–959.Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/10835412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queirós, J. , Gortázar, C. , & Alves, P. C. (2020). Deciphering anthropogenic effects on the genetic background of the red deer in the Iberian Peninsula. Frontiers in Ecology and Evolution, 8, 147. 10.3389/fevo.2020.00147 [DOI] [Google Scholar]
- Randi, E. (2008). Detecting hybridization between wild species and their domesticated relatives. Molecular Ecology, 17(1), 285–293. 10.1111/j.1365-294X.2007.03417.x [DOI] [PubMed] [Google Scholar]
- Ravinet, M. , Faria, R. , Butlin, R. K. , Galindo, J. , Bierne, N. , Rafajlović, M. , Noor, M. A. F. , Mehlig, B. , & Westram, A. M. (2017). Interpreting the genomic landscape of speciation: A road map for finding barriers to gene flow. Journal of Evolutionary Biology, 30(8), 1450–1477. [DOI] [PubMed] [Google Scholar]
- Reid, K. , Carlos Garza, J. , Gephard, S. R. , Caccone, A. , Post, D. M. , & Palkovacs, E. P. (2020). Restoration‐mediated secondary contact leads to introgression of alewife ecotypes separated by a colonial‐era dam. Evolutionary Applications, 13(4), 652–664. 10.1111/eva.12890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhymer, J. M. , & Simberloff, D. (1996). Extinction by hybridization and introgression. Annual Review of Ecology and Systematics, 27(1), 83–109. 10.1146/annurev.ecolsys.27.1.83 [DOI] [Google Scholar]
- Ruddiman, W. F. (2003). The anthropogenic greenhouse era began thousands of years ago. Climatic Change, 61(3), 261–293. 10.1023/B:CLIM.0000004577.17928.fa [DOI] [Google Scholar]
- Rudolfsen, T. , Ruppert, J. L. W. , Taylor, E. B. , Davis, C. S. , Watkinson, D. A. , & Poesch, M. S. (2019). Habitat use and hybridisation between the Rocky Mountain sculpin (Cottus sp.) and slimy sculpin (Cottus cognatus). Freshwater Biology, 64(3), 391–404. 10.1111/fwb.13225 [DOI] [Google Scholar]
- Runemark, A. , Vallejo‐Marin, M. , & Meier, J. I. (2019). Eukaryote hybrid genomes. PLoS Genetics, 15(11), e1008404. 10.1371/journal.pgen.1008404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford, S. , Rossetto, M. , Bragg, J. G. , McPherson, H. , Benson, D. , Bonser, S. P. , & Wilson, P. G. (2018). Speciation in the presence of gene flow: Population genomics of closely related and diverging Eucalyptus species. Heredity, 1, 10.1038/s41437-018-0073-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltonstall, K. , Lambert, A. M. , & Rice, N. (2016). What happens in Vegas, better stay in Vegas: Phragmites australis hybrids in the Las Vegas Wash. Biological Invasions, 18(9), 2463–2474. 10.1007/s10530-016-1167-5 [DOI] [Google Scholar]
- Salvatori, V. , Godinho, R. , Braschi, C. , Boitani, L. , & Ciucci, P. (2019). High levels of recent wolf × dog introgressive hybridization in agricultural landscapes of central Italy. European Journal of Wildlife Research, 65(5), 1–14. 10.1007/s10344-019-1313-3 [DOI] [Google Scholar]
- Schulte, U. , Veith, M. , & Hockkirch, A. (2012). Rapid genetic assimilation of native wall lizard populations (Podarcis muralis) through extensive hybridization with introduced lineages. Molecular Ecology, 21(17), 4313–4326. 10.1111/j.1365-294X.2012.05693.x [DOI] [PubMed] [Google Scholar]
- Schwenk, K. , Brede, N. , & Streit, B. (2008). Introduction. Extent, processes and evolutionary impact of interspecific hybridization in animals. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 363(1505), 2805–2811. 10.1098/rstb.2008.0055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, P. A. , Glenn, T. C. , & Rissler, L. J. (2019). Formation of a recent hybrid zone offers insight into the geographic puzzle and maintenance of species boundaries in musk turtles. Molecular Ecology, 28(4), 761–771. 10.1111/mec.14983 [DOI] [PubMed] [Google Scholar]
- Seehausen, O. (2004). Hybridization and adaptive radiation. Trends in Ecology and Evolution, 19(4), 198–207. 10.1016/j.tree.2004.01.003 [DOI] [PubMed] [Google Scholar]
- Seehausen, O. , Takimoto, G. , Roy, D. , & Jokela, J. (2008). Speciation reversal and biodiversity dynamics with hybridization in changing environments. Molecular Ecology, 17(1), 30–44. 10.1111/j.1365-294X.2007.03529.x [DOI] [PubMed] [Google Scholar]
- Seehausen, O. , Van Alphen, J. J. M. , & Witte, F. (1997). Cichlid fish diversity threatened by eutrophication that curbs sexual selection. Science, 277(5333), 1808–1811. 10.1126/science.277.5333.1808 [DOI] [Google Scholar]
- Sefc, K. M. , Mattersdorfer, K. , Hermann, C. M. , & Koblmüller, S. (2017). Past lake shore dynamics explain present pattern of unidirectional introgression across a habitat barrier. Hydrobiologia, 791(1), 69–82. 10.1007/s10750-016-2791-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendell‐Price, A. T. , Ruegg, K. C. , Anderson, E. C. , Quilodrán, C. S. , Van Doren, B. M. , Underwood, V. L. , Coulson, T. , & Clegg, S. M. (2020). The genomic landscape of divergence across the speciation continuum in island‐colonising silvereyes (Zosterops lateralis). G3: Genes, Genomes, Genetics, 10(9), 3147–3163. 10.1534/g3.120.401352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setter, D. , Mousset, S. , Cheng, X. , Nielsen, R. , DeGiorgio, M. , & Hermisson, J. (2020). VolcanoFinder: Genomic scans for adaptive introgression. PLoS Genetics, 16(6), e1008867. 10.1371/journal.pgen.1008867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyoum, S. , Adams, D. H. , Matheson, R. E. , Whittington, J. A. , Alvarez, A. C. , Sheridan, N. E. , Panzner, K. , & Puchulutegui, C. (2020). Genetic relationships and hybridization among three western Atlantic sparid species: Sheepshead (Archosargus probatocephalus), sea bream (A. rhomboidalis) and pinfish (Lagodon rhomboides). Conservation Genetics, 21(1), 161–173. 10.1007/s10592-019-01244-7 [DOI] [Google Scholar]
- Sivyer, L. , Morgan‐Richards, M. , Koot, E. , & Trewick, S. A. (2018). Anthropogenic cause of range shifts and gene flow between two grasshopper species revealed by environmental modelling, geometric morphometrics and population genetics. Insect Conservation and Diversity, 11(5), 415–434. 10.1111/icad.12289 [DOI] [Google Scholar]
- Slabbekoorn, H. , Bouton, N. , van Opzeeland, I. , Coers, A. , ten Cate, C. , & Popper, A. N. (2010). A noisy spring: The impact of globally rising underwater sound levels on fish. Trends in Ecology and Evolution, 25(7), 419–427. 10.1016/j.tree.2010.04.005 [DOI] [PubMed] [Google Scholar]
- Slabbekoorn, H. , & Ripmeester, E. A. P. (2008). Birdsong and anthropogenic noise: Implications and applications for conservation. Molecular Ecology, 17(1), 72–83. 10.1111/j.1365-294X.2007.03487.x [DOI] [PubMed] [Google Scholar]
- Smadja, C. , & Butlin, R. K. (2009). On the scent of speciation: The chemosensory system and its role in premating isolation. Heredity, 102(1), 77–97. 10.1038/hdy.2008.55 [DOI] [PubMed] [Google Scholar]
- Song, Y. , Endepols, S. , Klemann, N. , Richter, D. , Matuschka, F.‐R. , Shih, C.‐H. , Nachman, M. W. , & Kohn, M. H. (2011). Adaptive introgression of anticoagulant rodent poison resistance by hybridization between old world mice. Current Biology, 21(15), 1296–1301. 10.1016/J.CUB.2011.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen, W. , Broadgate, W. , Deutsch, L. , Gaffney, O. , & Ludwig, C. (2015). The trajectory of the Anthropocene: The great acceleration. The Anthropocene Review, 2(1), 81–98. 10.1177/2053019614564785 [DOI] [Google Scholar]
- Stukenbrock, E. H. (2016). The role of hybridization in the evolution and emergence of new fungal plant pathogens. Phytopathology, 106(2), 104–112. 10.1094/PHYTO-08-15-0184-RVW [DOI] [PubMed] [Google Scholar]
- Taillebois, L. , Sabatino, S. , Manicki, A. , Daverat, F. , Nachón, D. J. , & Lepais, O. (2020). Variable outcomes of hybridization between declining Alosa alosa and Alosa fallax . Evolutionary Applications, 13(4), 636–651. 10.1111/eva.12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, S. A. , & Larson, E. L. (2019). Insights from genomes into the evolutionary importance and prevalence of hybridization in nature. Nature Ecology & Evolution, 3(2), 170–177. 10.1038/s41559-018-0777-y [DOI] [PubMed] [Google Scholar]
- Taylor, S. A. , Larson, E. L. , & Harrison, R. G. (2015). Hybrid zones: Windows on climate change. Trends in Ecology and Evolution, 30(7), 398–406. 10.1016/j.tree.2015.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibihika, P. D. , Curto, M. , Alemayehu, E. , Waidbacher, H. , Masembe, C. , Akoll, P. , & Meimberg, H. (2020). Molecular genetic diversity and differentiation of Nile tilapia (Oreochromis niloticus, L. 1758) in East African natural and stocked populations. BMC Evolutionary Biology, 20(1), 16. 10.1186/s12862-020-1583-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiesmeyer, A. , Ramos, L. , Manuel Lucas, J. , Steyer, K. , Alves, P. C. , Astaras, C. , Brix, M. , Cragnolini, M. , Domokos, C. , Hegyeli, Z. , Janssen, R. , Kitchener, A. C. , Lambinet, C. , Mestdagh, X. , Migli, D. , Monterroso, P. , Mulder, J. L. , Schockert, V. , Youlatos, D. , … Nowak, C. (2020). Range‐wide patterns of human‐mediated hybridisation in European wildcats. Conservation Genetics, 21(2), 247–260. 10.1007/s10592-019-01247-4 [DOI] [Google Scholar]
- Todesco, M. , Pascual, M. A. , Owens, G. L. , Ostevik, K. L. , Moyers, B. T. , Hübner, S. , Heredia, S. M. , Hahn, M. A. , Caseys, C. , Bock, D. G. , & Rieseberg, L. H. (2016). Hybridization and extinction. Evolutionary Applications, 9(7), 892–908. 10.1111/eva.12367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia‐Montoya, W. A. , Elfekih, S. , North, H. L. , Meier, J. I. , Warren, I. A. , Tay, W. T. , Gordon, K. H. J. , Specht, A. , Paula‐Moraes, S. V. , Rane, R. , Walsh, T. K. , & Jiggins, C. D. (2020). Adaptive introgression across semipermeable species boundaries between local Helicoverpa zea and invasive Helicoverpa armigera moths. Molecular Biology and Evolution, 37(9), 2568–2583. 10.1093/molbev/msaa108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo‐Marín, M. , & Hiscock, S. J. (2016). Hybridization and hybrid speciation under global change. The New Phytologist, 211(4), 1170–1187. 10.1111/nph.14004 [DOI] [PubMed] [Google Scholar]
- van Hengstum, T. , Lachmuth, S. , Oostermeijer, J. G. B. , den Nijs, J. C. M. , Meirmans, P. G. , & van Tienderen, P. H. (2012). Human‐induced hybridization among congeneric endemic plants on Tenerife, Canary Islands. Plant Systematics and Evolution, 298(6), 1119–1131. 10.1007/s00606-012-0624-6 [DOI] [Google Scholar]
- van Wyk, A. M. , Dalton, D. L. , Hoban, S. , Bruford, M. W. , Russo, I.‐R.‐M. , Birss, C. , Grobler, P. , van Vuuren, B. J. , & Kotzé, A. (2017). Quantitative evaluation of hybridization and the impact on biodiversity conservation. Ecology and Evolution, 7(1), 320–330. 10.1002/ece3.2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wyk, A. M. , Dalton, D. L. , Kotzé, A. , Grobler, J. P. , Mokgokong, P. S. , Kropff, A. S. , & Jansen van Vuuren, B. (2019). Assessing introgressive hybridization in roan antelope (Hippotragus equinus): Lessons from South Africa. PLoS One, 14(10), e0213961. 10.1371/journal.pone.0213961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux, F. , Bohn, S. , Hyde, J. R. , & O’Malley, K. G. (2021). Adaptive markers distinguish North and South Pacific Albacore amid low population differentiation. Evolutionary Applications, eva.13202. 10.1111/eva.13202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- vonHoldt, B. M. , Brzeski, K. E. , Wilcove, D. S. , & Rutledge, L. Y. (2018). Redefining the Role of Admixture and Genomics in Species Conservation. Conservation Letters, 11(2), e12371. 10.1111/conl.12371 [DOI] [Google Scholar]
- Vonlanthen, P. , Bittner, D. , Hudson, A. G. , Young, K. A. , Müller, R. , Lundsgaard‐Hansen, B. , Roy, D. , Di Piazza, S. , Largiader, C. R. , & Seehausen, O. (2012). Eutrophication causes speciation reversal in whitefish adaptive radiations. Nature, 482(7385), 357–362. 10.1038/nature10824 [DOI] [PubMed] [Google Scholar]
- Waters, C. N. , Zalasiewicz, J. , Summerhayes, C. , Barnosky, A. D. , Poirier, C. , Ga uszka, A. , Cearreta, A. , Edgeworth, M. , Ellis, E. C. , Ellis, M. , Jeandel, C. , Leinfelder, R. , McNeill, J. R. , Richter, D. D. , Steffen, W. , Syvitski, J. , Vidas, D. , Wagreich, M. , Williams, M. , … Wolfe, A. P. (2016). The Anthropocene is functionally and stratigraphically distinct from the Holocene. Science, 351(6269), aad2622. 10.1126/science.aad2622 [DOI] [PubMed] [Google Scholar]
- Weeks, A. R. , Heinze, D. , Perrin, L. , Stoklosa, J. , Hoffmann, A. A. , Van Rooyen, A. , Kelly, T. , & Mansergh, I. (2017). Genetic rescue increases fitness and aids rapid recovery of an endangered marsupial population. Nature Communications, 8(1), 1–6. 10.1038/s41467-017-01182-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley, A. R. , Fitzpatrick, S. W. , Funk, W. C. , & Tallmon, D. A. (2015). Genetic rescue to the rescue. Trends in Ecology and Evolution, 30(1), 42–49. 10.1016/j.tree.2014.10.009 [DOI] [PubMed] [Google Scholar]
- Whitney, K. D. , Randell, R. A. , & Rieseberg, L. H. (2006). Adaptive introgression of herbivore resistance traits in the weedy sunflower Helianthus annuus . The American Naturalist, 167(6), 794–807. 10.1086/504606 [DOI] [PubMed] [Google Scholar]
- Wielstra, B. , Burke, T. , Butlin, R. K. , Schaap, O. , Shaffer, H. B. , Vrieling, K. , & Arntzen, J. W. (2016). Efficient screening for ‘genetic pollution’ in an anthropogenic crested newt hybrid zone. Conservation Genetics Resources, 8(4), 553–560. 10.1007/s12686-016-0582-3 [DOI] [Google Scholar]
- Willoughby, J. R. , Fernandez, N. B. , Lamb, M. C. , Ivy, J. A. , Lacy, R. C. , & DeWoody, J. A. (2015). The impacts of inbreeding, drift and selection on genetic diversity in captive breeding populations. Molecular Ecology, 24(1), 98–110. 10.1111/mec.13020 [DOI] [PubMed] [Google Scholar]
- Wolf, D. E. , Takebayashi, N. , & Rieseberg, L. H. (2001). Predicting the risk of extinction through hybridization. Conservation Biology, 15(4), 1039–1053. 10.1046/j.1523-1739.2001.0150041039.x [DOI] [Google Scholar]
- Wolf, J. B. W. , & Ellegren, H. (2017). Making sense of genomic islands of differentiation in light of speciation. Nature Reviews Genetics, 18(2), 87–100. [DOI] [PubMed] [Google Scholar]
- Wooten, J. A. , Sullivan, B. K. , Klooster, M. R. , Schwaner, T. D. , Sullivan, K. O. , Brown, A. D. , Takahashi, M. , & Bradford, P. R. (2019). Thirty years of hybridization between toads along the Agua Fria River in Arizona: Part II: Fine‐scale assessment of genetic changes over time using microsatellites. Journal of Herpetology, 53(2), 104. 10.1670/18-101 [DOI] [Google Scholar]
- Wu, C.‐I. (2001). The genic view of the process of speciation. Journal of Evolutionary Biology, 14(6), 851–865. [Google Scholar]
- Zhang, L. , Thibert‐Plante, X. , Ripa, J. , Svanbäck, R. , & Brännström, Å. (2019). Biodiversity loss through speciation collapse: Mechanisms, warning signals, and possible rescue. Evolution, 73(8), 1504–1516. 10.1111/evo.13736 [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Kim, B. , Lohmueller, K. E. , & Huerta‐Sánchez, E. (2020). The impact of recessive deleterious variation on signals of adaptive introgression in human populations. Genetics, 215(3), 799–812. 10.1534/genetics.120.303081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X. , Lutteropp, S. , Czech, L. , Stamatakis, A. , Looz, M. V. , & Rokas, A. (2020). Quartet‐based computations of internode certainty provide robust measures of phylogenetic incongruence. Systematic Biology, 69(2), 308–324. 10.1093/sysbio/syz058 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There are not data associated with this manuscript, which is solely based on literature.


