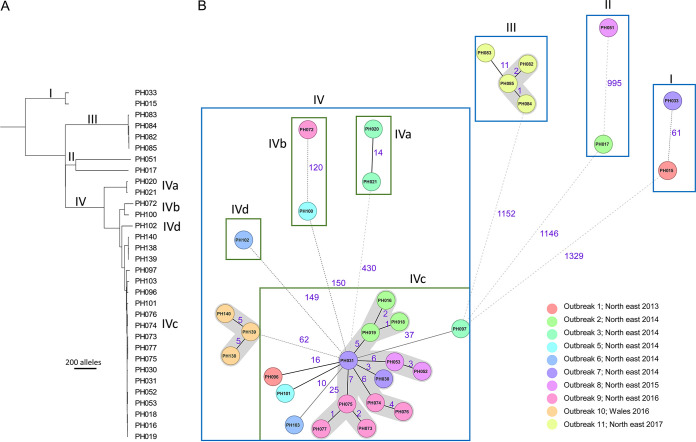
FIG 5.
Investigation of 32 Clostridium perfringens strains from outbreaks in care homes in the United Kingdom. (A) Neighbor-joining tree of the 32 C. perfringens strains using the cgMLST allele typing system, with lineages identified based on SNP analysis highlighted as in the original study (17). (B) Minimum spanning tree using cgMLST of the same genomes. Lineages are indicated with boxes, and outbreaks are color-coded as in the legend. Clusters were identified based on fewer than seven allelic mismatches (gray shading). Numbers on the connecting lines illustrate the number of target genes with different alleles, represented by solid lines for allele differences below 40 and dotted lines for allele differences above 40. The minimum spanning tree was generated using Ridom SeqSphere v07 (https://www.ridom.de/).