FIG 1.
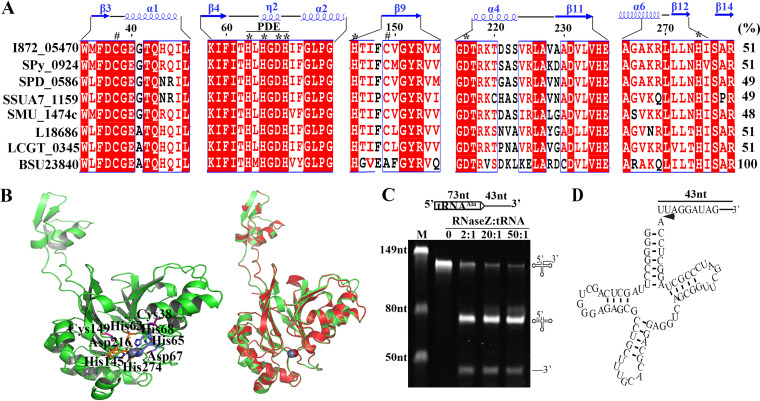
The S. oligofermentans So-RNaseZ processes the 3′ ends of tRNA precursors for mutation. (A) Sequence alignment of So-RNaseZ and the orthologs from Streptococcus, Lactococcus, and B. subtilis. Protein sequences were retrieved from the KEGG database and aligned using ClustalW. I872_05470: S. oligofermentans; Spy_0924: S. pyogenes; SPD_0586: S. pneumoniae; SSUA7_1159: S. suis; SMU_1474c: S. mutans; L18686: Lactococcus lactis; LCGT_0345: L. garvieae; and BSU23840: B. subtilis. The PDE domain, conserved amino acid residues binding Zn2+ (*), and cysteine residues at positions 38 and 149 (#) are indicated. The secondary structure of B. subtilis RNase Z and the So-RNaseZ amino acid residue numbers are shown at the top. The rightmost numbers represent the amino acid identities (%) of respective protein with BSU23840. (B) Homology modeling of the So-RNaseZ structure was constructed using SWISSMODEL by automatically selecting the B. subtilis RNase Z (4GCW) as a template. Conserved cysteine residues and amino acid residues for Zn2+ (sphere) binding are shown in the left panel, and superimposition of the So-RNaseZ homology model over 4GCW is shown in the right panel. (C) The 116 nt tRNAAla (I872_t10692) precursor RNA (schematic at top) was produced via in vitro transcription, and 50 ng of tRNA precursor was mixed with a gradient of increasing So-RNaseZ protein at indicated molar ratios. Detailed procedures are described in Materials and Methods. The endonucleolytic products were examined on 10% urea-PAGE gels. M, RNA marker with molecular weights indicated on the gel left; the precursor and cleaved product symbols are shown on the gel right. (D) Schematic of the tRNAAla precursor showing 5′ and 3′ RACE-determined So-RNaseZ cleavage sites located immediately downstream of the discriminator nucleotide and indicated by an arrow.