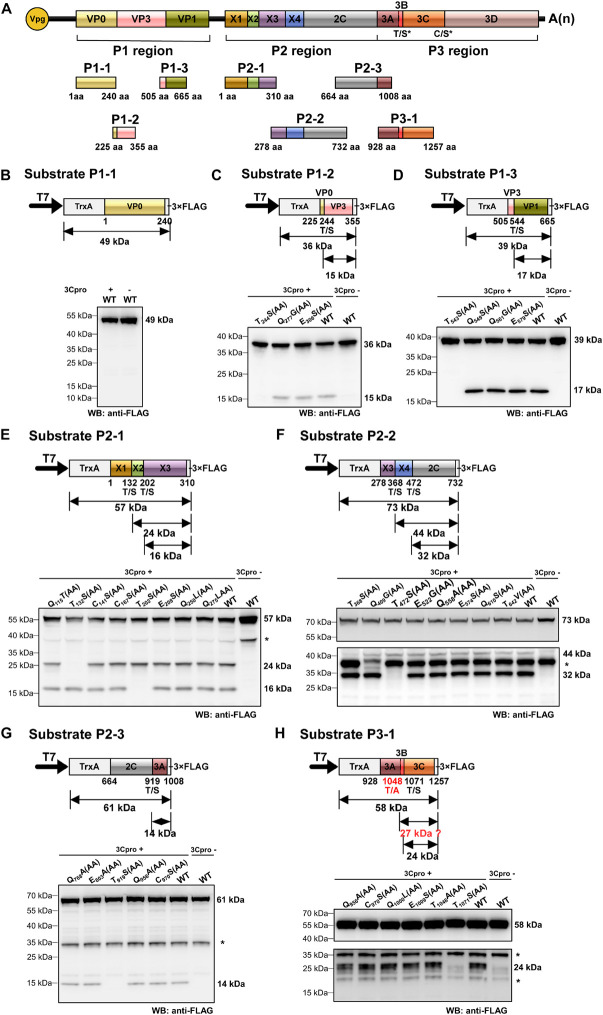
FIGURE 4.
Identification of cleavage sites in rice curl dwarf-associated picornavirus (RCDaV) polyproteins using in vitro trans-cleavage assay. (A) Schematic representation of protein segments from the three regions of RCDaV. (B–D) In vitro trans-cleavage analyses using the P1-1, P1-2, and P1-3 protein segments from P1 region as the substrates. (E–G) In vitro trans-cleavage analyses using the P2-1, P2-2, and P2-3 protein segments from P2 region as the substrates. (H) In vitro trans-cleavage analysis using the P3-1 protein segment from P3 region as the substrate. Site-directed mutants of the predicted cleavage sites were also used in individual assays. The calculated molecular mass of each protein segment is shown on the right side of the figure. The asterisks in (E–H) indicate non-specific protein bands. The lower panel blots in (F,H) were exposed for longer time to show the bands. Predicted T/A cleavage site is shown with a dashed line in (H), since processing was not detected at this site in the in vitro assays. Numbers below the boxes indicate amino acids positions.