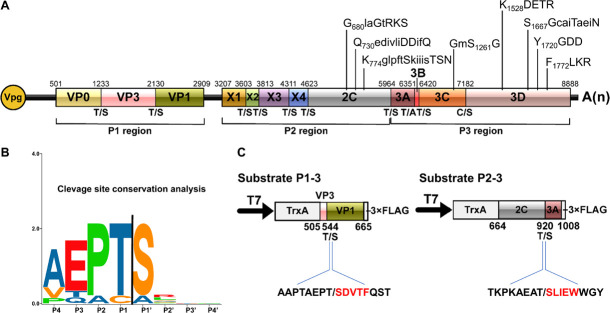
FIGURE 5.
Conservation analysis of cleavage sites in rice curl dwarf-associated picornavirus (RCDaV) polyproteins. (A) Schematic representation of the elaborate genome organization. The genome maps and each gene boxes were drawn to scale. Nucleotide position of individual protein is indicated above each gene box. The structural proteins are named as reported previously (Roberts and Groppelli, 2009). The cleavage sites are indicated at the borders between the two cleaved proteins. The conserved picornaviral motifs are indicated above the diagram. (B) Sequence logos of RCDaV cleavage sites. The P4–P1/the P1′–P4′ aa of cleavage sites are shown. Amino acids are color-coded according to their physicochemical characteristics. Neutral and polar, green; basic, blue; acidic, red; neutral and hydrophobic, black. Amino acids are shown as one-letter standard code. (C) The first five amino acids (red) in cleavage products of substrates P1-3 and P2-3 were determined through N-terminal Edman degradation sequencing. Slashes indicate the cleavage sites.